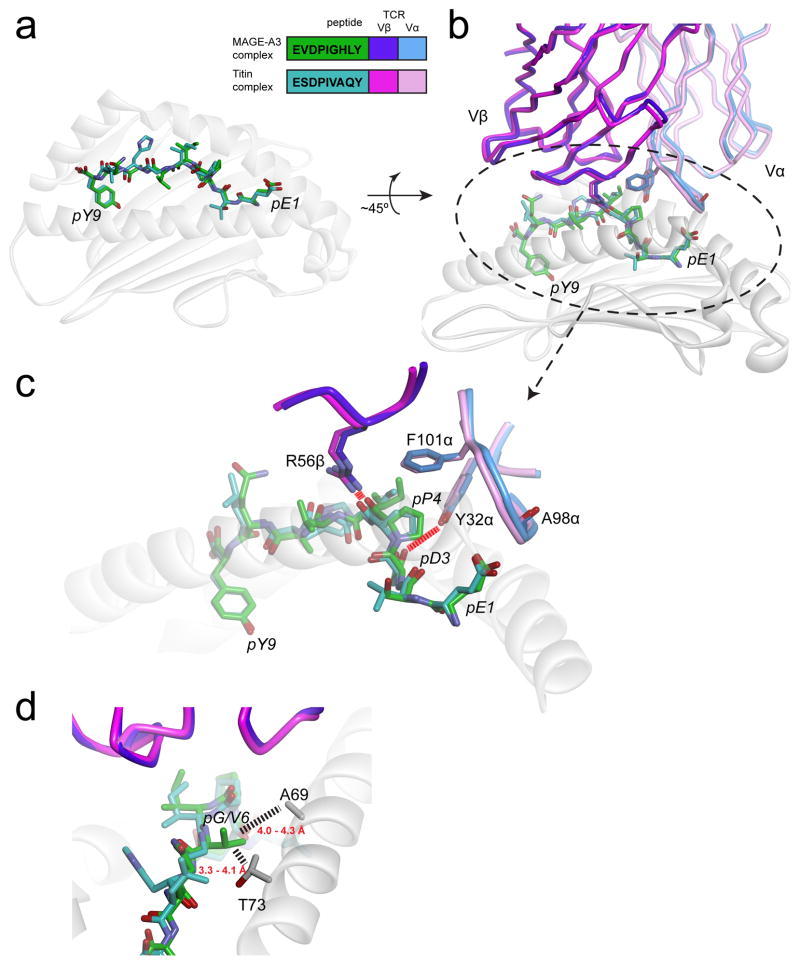
Figure 4.
The MAGE-A3 and Titin epitopes are presented and recognized almost identically by the MAG-IC3 TCR (74). A) Near-identical presentation of the two epitopes in the HLA-A1 peptide binding groove. The inset shows the peptide sequences and the color scheme used for all panels. B) Overview of how the two ligands are engaged by the MAG-IC3 TCR. C) Details of identical, key interactions in the two interfaces. Charge complementarity with pGlu1 is optimized by the positioning of the carbonyl oxygen of Ala98 of CDR3α away from the glutamate side chain (not shown is a salt-bridge from Arg170 of HLA-A1 that helps fix the glutamate). Tyr32 of CDR1α hydrogen bonds with pAsp3. Phe101 of CDR3α “caps” the hydrophobic pPro4, and Arg56 of CDR2β hydrogen bonds with the pPro4 backbone. Not evident in the figure is how pIle5 packs between the TCR and the HLA-A2 α2 helix. D) The Titin and MAGE-A3 peptides have a valine and a glycine at P6, respectively. Substitution with larger amino acids would result in clashes with Ala69 and Thr73 of the HLA-A1 α1 helix, explaining why pathogen-derived peptides similar to the Titin and MAGE-A3 peptides would not be recognized. Dashed lines show distances between the side chains of Val6 of the Titin peptide and residues of HLA-A1 (note that in generating this figure, the CDR1α loop in the two structures was optimized to better fit the experimental electron density and amino acid geometry, yielding coordinates slightly altered from those deposited in the PDB).