Figure 4. Ancient genomes provide evidence of natural selection in present-day southern African San populations.
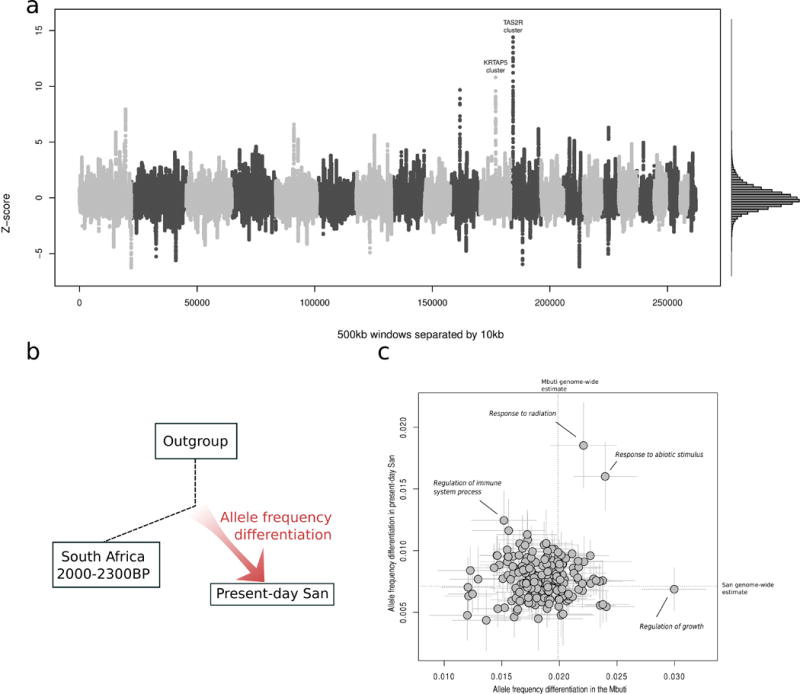
We computed branch-specific allele frequency differentiation in 6 present-day high-coverage San genomes compared to a pool of two ~2000 BP South African genomes as an outgroup, using two approaches. A) We computed the statistic in windows of 500 kb separated by 10 kb. We also estimated genome-wide average and standard deviation of the statistic using windows separated by at least 5 Mb, and transformed the genome wide distribution of the sliding windows to be approximately normal (right panel). We observe outliers 15 standard deviations from the mean in a taste receptor gene cluster on chromosome 12, and a secondary peak in the Keratin Associated Protein 4 gene cluster. The outgroup used was 4 Central African Mbuti genomes. See Table 2 for details on all major outlying regions. B) Illustration of the branch-specific allele frequency differentiation approach. C) We computed the statistic and block jackknife standard errors for 208 gene ontology categories with at least 50 genes each (y-axis). The outgroup used was western Africans. As a control to confirm that outlier categories do not show larger magnitudes of allele frequency differentiation across populations, we replaced the present-day San with the central African Mbuti (x-axis).
