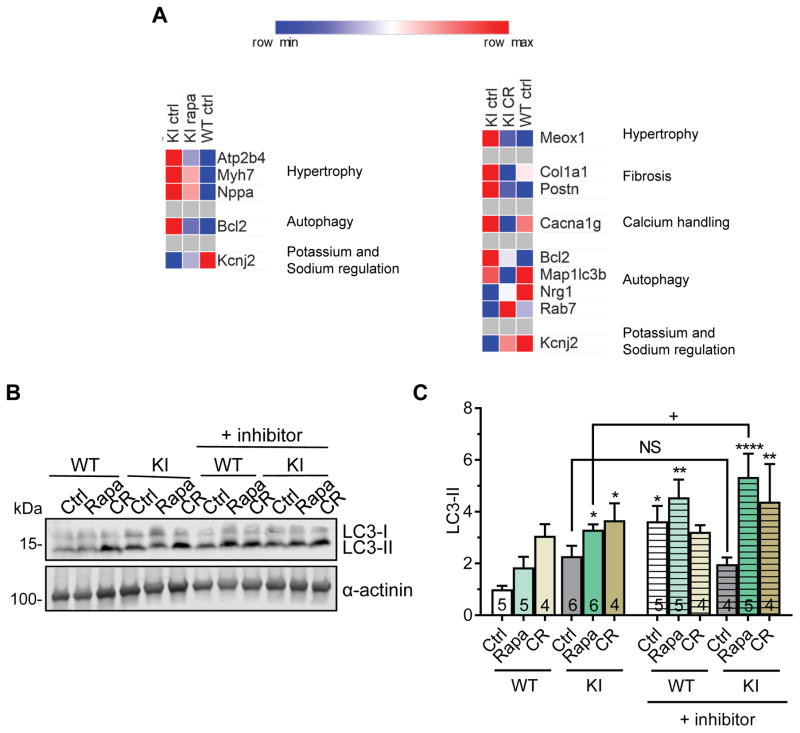
Figure 7. Gene expression analysis and autophagic flux in rapamycin-treated or calorie-restricted KI and WT mice.
KI and WT mice were treated for 9 weeks with either 2.24 mg/kg rapamycin (rapa), 40% caloric restriction (CR) or control treatment (ctrl). A, Heatmap of selected genes (threshold <0.8 or >1.2 fold change to KI ctrl) comparing gene expression of hypertrophy, fibrosis, calcium handling, autophagy and potassium and sodium regulation between KI ctrl, KI rapa or KI CR and WT ctrl mice. B, Representative Western blots of indicated proteins from mouse ventricular protein extracts (membrane-enriched fraction). α-actinin was used as loading control. C, LC3-II quantification (normalized to α-actinin) related to WT ctrl. Data are expressed as mean + s.e.m. with *P<0.05, **P<0.01 and ****P<0.0001 vs. WT ctrl, one-way ANOVA plus Dunnett’s post-test, and non-significant (NS) and +P<0.05 vs. indicated group (comparing with and without inhibitor), unpaired Student’s t-test. Number of animals is indicated in the bars.