Fig 1. Computational analysis of DinGNm.
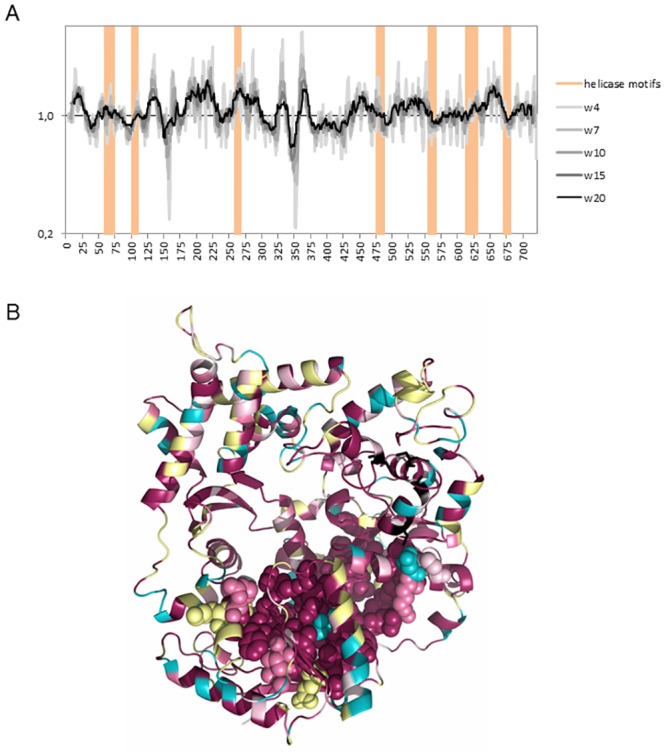
A) Plotting of the similarity of DinGNm sequences from PubMLST with sliding window sizes (w) ranging from 4 to 20 aa. Data were generated with plotcon (EMBOSS) and the average similarity for the whole sequence was set to 1. The helicase motive I, Ia, II, III, IV, V, and VI are under laid in orange colour. B) Similarity of the DinGNm sequences taken from PubMLST plotted onto the predicted structure of DinGNm from MC58 using default Consurf colouring with cysteins shown in black. The helicase motifs are shown as spheres and the rest of the protein is shown as cartoon.
