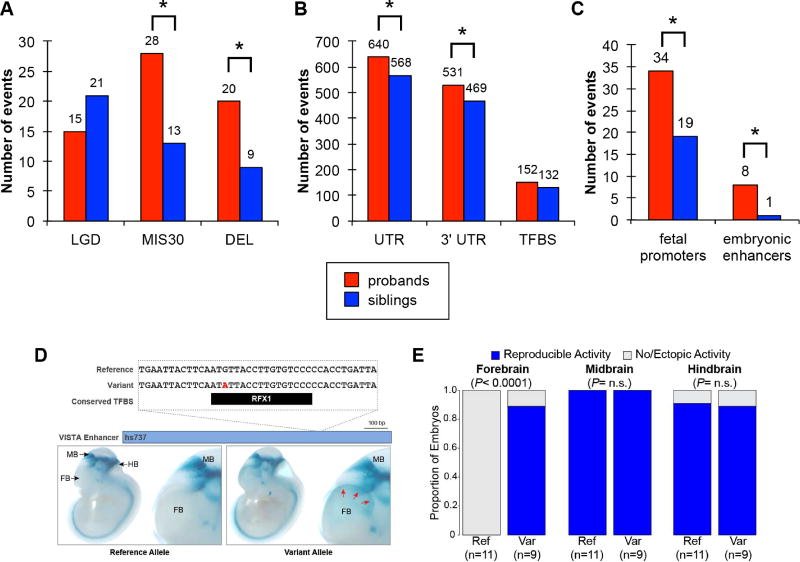
Figure 2. Proband–sibling DNM differences by functional annotation.
Number of autosomal de novo variants by functional category by (A) coding variants (LGD=likely gene-disrupting, MIS30=missense with CADD score >30, DEL=exonic deletion); (B) putative noncoding regulatory variants (UTR=untranslated region, 3' UTR=3' untranslated region, TFBS=putative noncoding regulatory with a transcription factor binding site); and (C) ENCODE/ChromHMM putative regulatory variants (fetal promoters=within a TFBS in a fetal brain transcription start site and embryonic enhancers=within a TFBS in a human embryonic stem cell strong enhancer). * Indicates nominal significance (p<0.05) by FET. (D) One of the three de novo sequence variants identified in autism probands that was tested for in vivo enhancer activity in the CNS (reference allele from enhancer.lbl.gov) (Visel et al., 2007). We extended the previous assessment for the reference allele in our current study by also testing the variant allele. For the locus, we show, from top to bottom, the human genome reference allele, the patient variant (red text), the location of a conserved TFBS near the variant, the VISTA enhancer with hs number (blue bar), and representative transgenic embryonic day 11.5 mouse embryos for the reference and variant alleles, respectively, displaying the enhancer activity pattern (blue staining). Whole embryos are shown on left, with enlarged images of the forebrain on the right. FB: forebrain, MB: midbrain, HB: hindbrain. (E) Results of the enhancer assay identified a novel forebrain enhancer activity pattern being driven by the allele containing the de novo patient variant. The expression in both the midbrain and hindbrain were unaffected. P-values by FET.