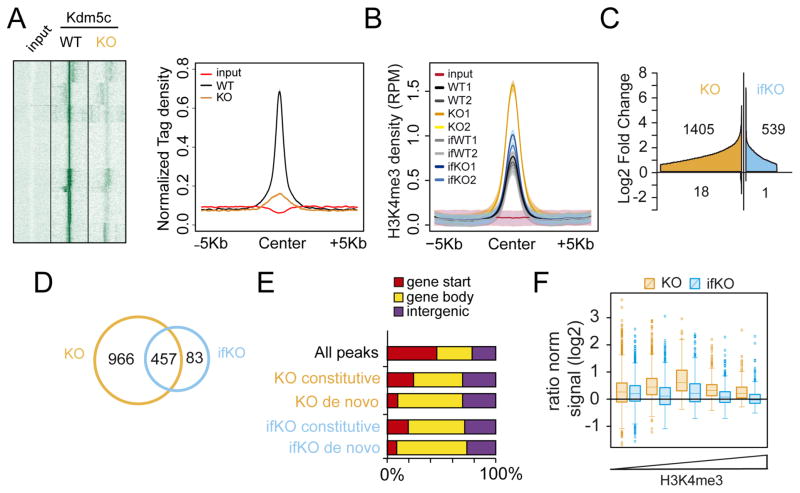
Figure 3. Lack of Kdm5c increases H3K4me3 levels throughout the neuronal epigenome.
A. Heatmap (right) and line plot (left) showing Kdm5c density at Kdm5c-enriched regions (as detected in hippocampal chromatin of wild type mice) in KO and control littermates. Color intensity in the heatmap is proportional to read density. B. Density plot showing the average H3K4me3 signal (expressed as reads per million, RPM) of DHMPs for WT, KO, ifWT and ifKO replicates. Input values are also shown. Shaded areas represent the 95% confidence interval (CI). C. Cumulative graph showing the number of DHMPs (BH p adj. < 0.05, LFC > 0.58) and their fold change with respect to the relative control (i.e., KO vs WT; ifKO vs ifWT). DHMPs were ordered by Log2FC and plotted as separate entities on the x-axis. D. Venn diagram depicting the overlap between the DHMPs of KO (vs WT) and ifKO (vs ifWT) mice. E. Distribution of DHMPs in function of gene feature, in Kdm5c-KOs and ifKOs. “Gene start” refers to peaks located between ±1 Kb of the first TSS; “Gene body” denotes internal peaks (including alternative TSSs); and “Intergenic” refers to peaks that are farther than −1 Kb from the TSS of any Refseq annotated gene. F. H3K4me3 changes at putative enhancers (intra- and intergenic H3K27ac-rich regions not overlapping with a TSS) in KOs and ifKOs. Enhancers were classified according to their H3K4me3 content (see Fig. S3G–H for additional detail).