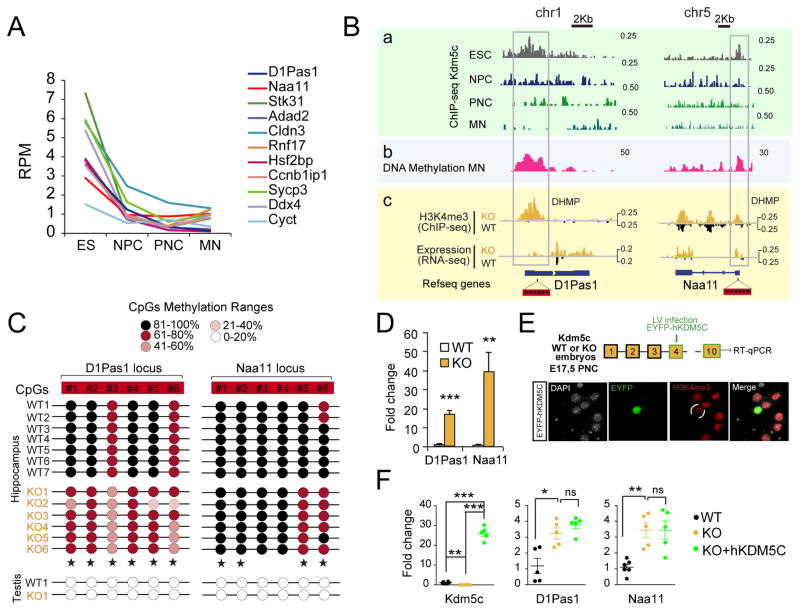
Figure 6. Germinal loss of Kdm5c prevents the silencing of germ line genes.
A. Developmental loss of Kdm5c occupancy at the TSS of germ line genes (pink dots in Fig 4E). RPM: reads per million. B. Genomic profiles for representative germ line genes. From top to bottom, we show (a) Kdm5c-ChIP-seq profiles in ESC, NPC (Outchkourov et al., 2013), PNC (Iwase et al., 2016), and adult brain (MN, this study); (b) MeDIP profiles in adult brain of wild type mice (Halder et al., 2016), and (c) RNAseq and H3K4me3-ChIPseq experiments in adult hippocampus of WT and KO mice (this study). C. DNA methylation at specific CpGs (red boxes in Fig. 6B) at the promoter of D1Pas1 and Naa11 in hippocampal chromatin of WT and KO littermates (star symbols label significant DNA methylation reduction in KOs). Testis DNA was used as control for un-methylated DNA. D. RT-qPCR assays confirm the expression of germ line genes in the hippocampi of KOs (D1Pas1: t10 = 8.53, p < 0.0001; Naa11: t10 = 3.90, p = 0.003). E. Scheme of the experiment (top). Functionality of the hKDM5C construct was confirmed by transient transfection and examination of H3K4me3 levels in EYFP-expressing neurons (bottom). F. RT-qPCR assay confirmed the overexpression of germ line genes in PNCs from Kdm5c−/y embryos, infected or not with a hKDM5C-expressing lentivirus (Kdm5c, WT vs KO: t8 = 4.64, p = 0.002, KO vs KO+hKDM5C: t7 = 15.57, p < 0.0001, WT vs KO+hKDM5C: t9 = 18.54, p < 0.0001; D1Pas1, WT vs KO: t8 = 3.49, p = 0.008; KO vs KO+hKDM5C: t8 = 1.27, p = 0.2; Naa11, WT vs KO: t9 = 5.05, p = 0.0007; KO vs KO+hKDM5C: t8 = 0.0007, p = 1.0). Data are expressed as mean + s.e.m. ns: non-significant; * = p < 0.05; ** = p < 0.005; *** = p < 0.0005 in Student’s t test.