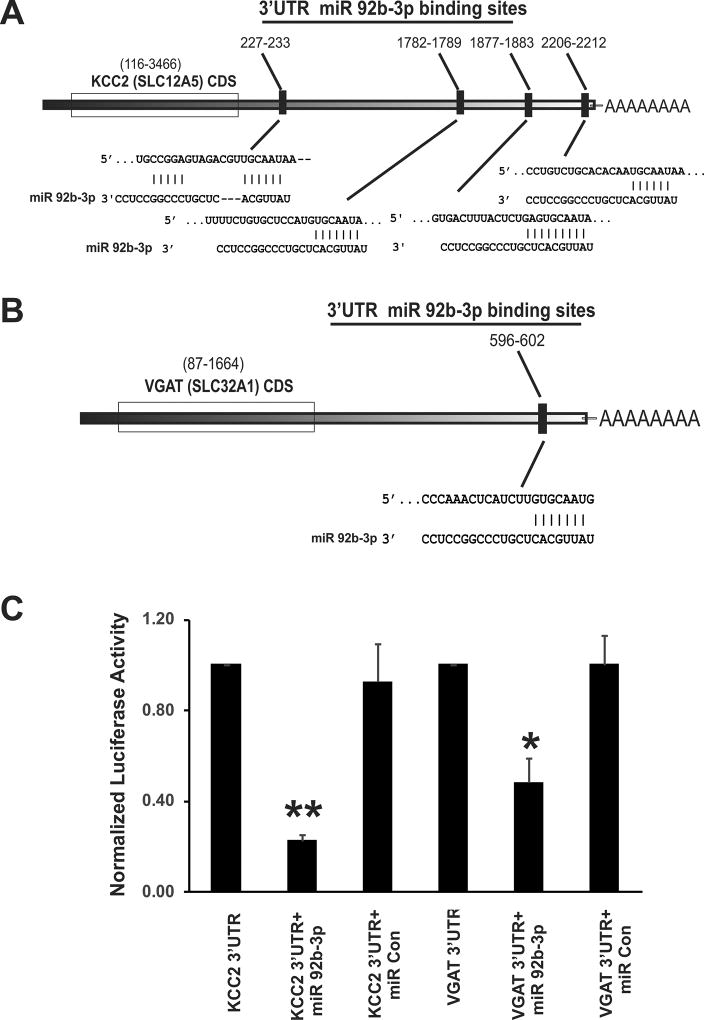
Figure 4.
Schematic representation of 3’ UTR of KCC2 and VGAT mRNA with complementary binding sequences for miR-92b-3p and Luciferase Assay validation. (A) 3’UTR of KCC2 (Slc12a5 gene) mRNA demonstrates four complimentary binding sequences (response elements) for the seed region of miR-92b-3p. (B) 3’UTR of VGAT (Slc32a1 gene) mRNA carries one complimentary binding site for miR-92b-3p. (C) miR-92b-3p interaction with 3’UTRs of KCC2 and VGAT in HEK293 cells. The percentage reduction in luciferase activity of the cells transfected with plasmid carrying either KCC2 or VGAT 3’UTRs and miR-92b-3p was calculated by considering 100% luciferase activity for cells transfected only with plasmid carrying the 3’UTR of the target gene. The data represent the normalized luciferase activity of three different sets of transfections. The percentage reduction in luciferase reduction activity of cells transfected with KCC2 3’UTR and miR 92b-3p mimic is significantly higher than cells transfected with KCC2 and miR-control mimic (fig. 4C, **p<0.001). Similarly, percentage reduction in luciferase activity of cells transfected with VGAT 3’UTR and miR 92b-3p is significantly higher than cells transfected with VGAT and miR control mimic (fig. 4C, *p<0.01).