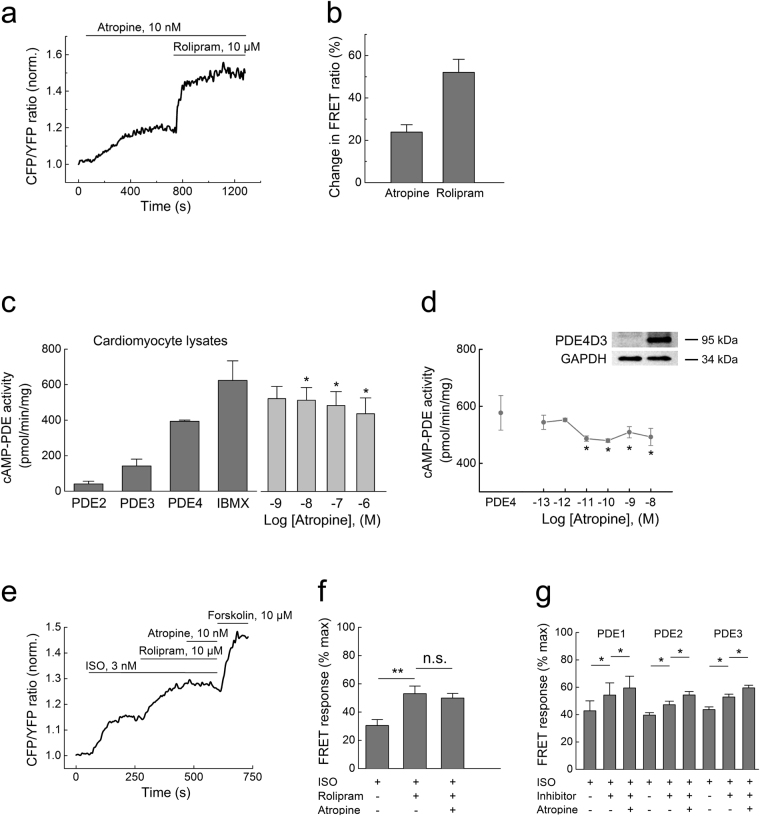
Figure 2.
Atropine inhibits PDE4 activity. (a) Single-cell FRET analysis of PDE4 inhibition in HEK293A cells expressing the Epac1-camps-PDE4 sensor. Cells were prestimulated with 1 µM ISO for 3 min before adding atropine (10 nM) and rolipram (10 µM) to pre-elevate intracellular cAMP levels. (b) Quantification of the data shown in a as a % change of the FRET ratio in response to atropine along with maximal effects measured by these sensors with respective inhibitors, mean ± s.e.m. (n = 6). Atropine inhibits cAMP-PDE activity in cardiomyocyte lysates (c) and recombinant PDE4D3 from transfected HEK293 cells (d), as measured by a classical in vitro PDE assay (n = 3–5). The basal PDE activity is represented by the “IBMX” bar in c or “PDE4” bar (rolipram-sensitive activity of PDE4D3 transfected minus vector-transfected control [Co] cell lysates, a representative immunoblot for PDE4D3 is shown) in (d.) *p < 0.05 compared to basal PDE activity by one-way ANOVA. (e) Preincubation of cardiomyocytes with 10 µM rolipram prevents the atropine (10 nM) mediated increase of cAMP after ISO stimulation (3 nM to avoid sensor saturation by rolipram). Representative experiment (n = 6), data analysis is in f. (g) preincubation of cells with 30 µM 8-MMX to block PDE1 (n = 6), 100 nM of the PDE2 inhibitor BAY 60–7550 (n = 6) or 10 µM of the PDE3 inhibitor cilostamide (n = 7) under the same experimental conditions (except for 100 nM ISO used to prestimulate cAMP levels) did not abolish the effect of atropine. *p < 0.05, **p < 0.01, n.s. – not significant by paired t-test.