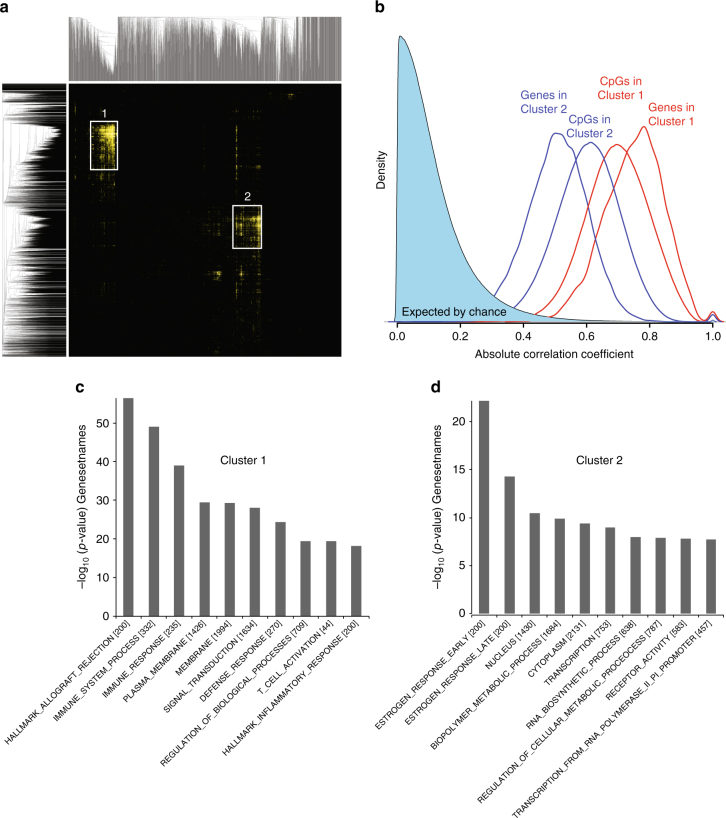
Fig. 1.
Identification of expression-methylation QTL (emQTL) a Unsupervised clustering of (−log) p-values of the emQTL by Pearson’s correlation and average linkage revealed two main clusters of CpG-gene pairs. Rows represent CpGs and columns represents genes. Yellow and light yellow spots show highly significant associations between CpG methylation and gene expression. b Density plot showing the degree of absolute co-expression of genes and co-methylation of CpGs in Cluster 1 (red) and Cluster 2 (blue). c, d Gene set enrichment analysis in Cluster 1 (c, n = 160) and Cluster 2 (d, n = 270) using MSigDB (H and C5 databases). The height of the bars represents the level of enrichment measured as a ratio between the number of genes overlapping an MSigDB H or C5 gene set over the expected frequency if such overlaps were to occur at random