ABSTRACT
It is only recently that the abundant presence of circular RNAs (circRNAs) in all kingdoms of Life, including the hyperthermophilic archaeon Pyrococcus abyssi, has emerged. This led us to investigate the physiologic significance of a previously observed weak intramolecular ligation activity of Pab1020 RNA ligase. Here we demonstrate that this enzyme, despite sharing significant sequence similarity with DNA ligases, is indeed an RNA-specific polynucleotide ligase efficiently acting on physiologically significant substrates. Using a combination of RNA immunoprecipitation assays and RNA-seq, our genome-wide studies revealed 133 individual circRNA loci in P. abyssi. The large majority of these loci interacted with Pab1020 in cells and circularization of selected C/D Box and 5S rRNA transcripts was confirmed biochemically. Altogether these studies revealed that Pab1020 is required for RNA circularization. Our results further suggest the functional speciation of an ancestral NTase domain and/or DNA ligase toward RNA ligase activity and prompt for further characterization of the widespread functions of circular RNAs in prokaryotes. Detailed insight into the cellular substrates of Pab1020 may facilitate the development of new biotechnological applications e.g. in ligation of preadenylated adaptors to RNA molecules.
KEYWORDS: Archaea, circular RNA, computational biology, RNA ligase, RNA-Seq
Introduction
For a long time, circular RNA molecules (circRNAs) lacking 3′ or 5′ termini were considered an unusual form of nucleic acids found in few viroids or viruses using a single-stranded RNA molecule as genetic material, participating in maturation of some tRNA genes, or, alternatively, a result of aberrant RNA splicing (for recent reviews, see1-3). However, numerous recent genome-wide experimental and computational studies (RNA-seq analyses) tailored toward detection of circRNAs have revealed this class of RNA molecules as abundant in eukarya, including humans. Evolutionary conservation of circRNAs has suggested that they are functionally important. Indeed, recent studies have revealed that circRNAs are differentially expressed in different human cell lines and tissues, serve as regulators of transcription and protein expression and can act as miRNA sponges.4-7 Several different types of RNA circles originating from diverse cellular processes have been identified. Most eukaryotic circRNAs result from splicing reactions that are catalyzed either by the spliceosome or ribozymes corresponding to Group I and Group II introns. The process called “backsplicing” that connects a downstream splice donor site (5′ splice site) to an upstream acceptor site (3′ splice site) is the most common mechanism producing circRNAs in eukaryotic cells. During the formation of circRNAs either 2′–5′ or 3′–5′ linkages have been detected.8,9 The resulting molecular “rings” or RNA circles resist degradation by exoribonucleases that require free termini and/or may have increased melting point in comparison to linear nucleic acid molecules. Circularization of RNA molecules may thus drastically influence the structure and/or shape of these molecules, presumably reflecting structural constraints brought about by circularization.
Although the majority of recent work on circRNAs has been performed using human cell lines, circular RNAs also exist in archaea and bacteria, in addition to the aforementioned viroids and RNA viruses. This raises the question how circular RNA molecules are formed in prokaryotes, where RNA splicing is a rare phenomenon. RNA-seq methodology revealed an abundant genome-wide presence of circRNAs in the transcriptome of the archaeon Sulfolobus solfataricus10 such as tRNA introns and rRNA processing intermediates, as well as several protein-coding genes, and many smaller non-coding RNAs (e.g., box C/D RNAs). This genome-wide study is also in agreement with earlier studies that have revealed the presence of circular forms in excised tRNA-introns,11,12 introns of rRNAs,13,14 rRNAs processing intermediates15 and box C/D RNAs in archaea.16 Note that box C/D RNAs guide site-specific modification (2′-0-methylation) of rRNA and small nuclear RNA in eukaryotes and tRNA in archaea.17 RNA-seq data obtained using the “minimal” archaeon Nanoarchaeum equitans also revealed circular box C/D RNAs sequences.18 The specific case where the cleaved-out intron contains a C/D box RNA that guide 2′-O-methylation on nucleotides in the anticodon loop of mature tRNA-Trp should also be highlighted.12,19 At least 2 separate enzymes, an endonuclease and a ligase, are known to be involved in pre-tRNA splicing in archaeal cell free extracts.20 The endonuclease cleaves in a characteristic bulge-helix-bulge structure (BHB) of pre-tRNA-Trp, producing 2′-3-cyclic phosphate and 5′-hydroxyl termini joined by the ligase.11 In addition to ligation of resulting tRNA halves, circularization of the pre-tRNA Trp intron occurs in a mechanistically similar ligation reaction, as observed using H. volcanii extracts.11,19
Considering the widespread presence of circRNAs in the third domain of life is now evident, further studies on archaeal RNA ligases are now necessary. Up to date, 2 evolutionary unrelated families of RNA ligases capable of joining single stranded RNA molecules have been identified in archaea. RtcB proteins seem to function in most, if not all, archaea as GTP-dependent tRNA-splicing ligases and join spliced tRNA molecules halves to form mature-sized tRNAs molecules e.g., in archaeal precursor tRNA-Trp.21 The observation that the genomes of some archaea contain 2 open reading frames, previously predicted to function as DNA ligases, led to the discovery of a putative second family of archaeal RNA ligases.22,23 The founding member of this putative RNA ligase family is the Pyrococcus abyssi reading frame Pab1020. This family (InterPro code IPR001072) currently contains 170 archaeal and 35 bacterial homologs but little is known concerning the molecular function of this conserved protein family. We have previously shown that, unexpectedly, Pab1020 catalyzed the circularization of physiologically non-significant oligoribonucleotides in an ATP-dependent reaction.22 Similar results have also been reported for closely related Methanobacterium protein that catalyzes the intramolecular ligation of 5′-P single-stranded RNA to form a covalently closed circular RNA molecule.24-27
Here using a combination of biochemical, RNA-seq and computational analyses, we have investigated the molecular function of the Pab1020 RNA ligase family.24,28 Our genome-wide RNA co-immunoprecipitation studies, using affinity purified Pab1020 antibodies, led to the discovery of a large number of circular RNAs that specifically interact with Pab1020 RNA ligase in P. abyssi cells and were indeed efficiently circularized by the Pab1020 in vitro. Our studies also indicate a widespread importance of circular RNAs in prokaryotes and suggest a functional speciation of an archaeal polynucleotide ligase toward RNA circularization activity in many thermophilic archaea and bacteria. The identification of physiologic substrates for the Pab1020 RNA ligase may also facilitate development of new biotechnological applications for this enzyme family currently commercialized for e.g., labeling of 3′ termini of RNA.
Results
Domain structure of the Pab1020 family RNA ligases
Pab1020 is the founding member of the conserved Rnl3 family of RNA ligases28 that are predominantly found in hyperthermophiles (archaea, bacteria) and halophiles. Each Pab1020 monomer consists of 4 domains: the amino-terminal (N-term), the catalytic nucleotide transferase (NTase), the dimerization (Dim) and the C-terminal (C-term) domains [22, Fig. S1A]. To address why these proteins are frequently annotated as “DNA” ligases, we have performed several sequence similarity searches indicating that the central NTase domain of Pab1020 is closely related to the CDC9 domain found in ATP-dependent DNA ligases [residues 66–235, E-value 1.21−5, CDD database29]. This domain carries the conserved nucleotide binding domain and corresponds to the minimal catalytic core of this family of enzymes. More sensitive HHpred searches based on the pairwise comparison of profile hidden Markov models (HMMs) also indicated that this domain is related to COG1793 [ATP-dependent DNA ligases, E-value = 7.6−23) and COG0272 [NAD-dependent DNA ligases, E-value = 2.1−14]. Note also that the NTase domain of Pab1020 is 26% identical and 37.8% similar with the corresponding domain from the experimentally validated P. abyssi DNA ligase Pab2002 and carries all the expected motifs for ATP-dependent polynucleotide ligases.22 Additional HHpred searches using the Pab1020 protein sequence as a query further indicated that, at the sequence level, this proteins is related to ATP or NAD+-dependent DNA ligases, as well as different families of RNA ligases, mRNA capping enzymes and RNA repair enzymes (Table 1). These results explain why members of the Rnl3 family are frequently misannotated as DNA ligases. An excellent example of this is the putative Aquifex aeolicus DNA ligase (PDB code 3qwu_A in Table 1). This bacterial protein is very likely an RNA ligase, as it contains a dimer interface that is only conserved among the Rnl3 family members in the PDB databank.
Table 1.
Level of sequence similarity of Pab1020 with different protein families carrying the nucleotidyltransferase (NTase) domain.
| Hit | PDB code (chain) | Description of the hita | E-value | number of aligned positions | Query range | HMM Template |
|---|---|---|---|---|---|---|
| HMM-HMM comparison against protein structure database | ||||||
| 1 | 2vug_A | Pab1020; RNA ligase | 5.6E-63 | 381 | 1–382 | 8–389 |
| 2 | 5d1p_A | ATP-dependent RNA ligase | 2.2E-51 | 366 | 9–380 | 7–378 |
| 3 | 3qwu_A | Putative DNA ligase | 1.0E-50 | 361 | 17–382 | 4–370 |
| 4 | 1dgs_A | DNA ligase; NAD-dependent | 1.3E-38 | 270 | 37–344 | 32–369 |
| 5 | 4 glx_A | DNA ligase, NAD-dependent | 6.0E-38 | 272 | 37–344 | 30–371 |
| 6 | 2owo_A | DNA ligase, NAD-dependent | 1.7E-37 | 272 | 37–344 | 30–371 |
| 7 | 3sgi_A | DNA ligase, NAD-dependent | 2.3E-38 | 272 | 37–344 | 39–381 |
| 8 | 1b04_A | DNA ligase, NAD-dependent | 2.1E-31 | 224 | 37–284 | 32–317 |
| 10 | 3jsl_A | DNA ligase; NAD+-dependent | 6.7E-30 | 219 | 37–260 | 30–310 |
| 14 | 5cot_A | Naegleria gruberi RNA ligase | 1.1E-16 | 173 | 62–244 | 133–330 |
| 15 | 1s68_A | RNA ligase 2; ATP-dependent | 5.7E-16 | 174 | 65–247 | 2–231 |
| 16 | 2hvq_A | T4 RNA Ligase 2, ATP-dependent | 1.0E-15 | 199 | 65–276 | 3–272 |
| 17 | 2c5u_A | RNA ligase, ATP-dependent | 2.8E-14 | 200 | 29–244 | 30–244 |
| 18 | 3l2p_A | DNA ligase 3, ATP-dependent | 1.6E-13 | 159 | 87–246 | 246–426 |
| 19 | 3w1b_A | DNA ligase 4, ATP-dependent | 2.3E-13 | 181 | 65–246 | 241–455 |
| 20 | 2cfm_A | Pyrococcus DNA ligase (ATP) | 2.9E-13 | 157 | 87–245 | 241–424 |
| 23 | 4pz7_A | mRNA-capping enzyme | 6.9E-14 | 202 | 43–259 | 21–259 |
| 27 | 1ckm_A | mRNA capping enzyme | 2.3E-12 | 165 | 66–252 | 54–245 |
| 28 | 1xdn_A | RNA editing ligase MP52 | 3.7E-12 | 173 | 65–244 | 8–263 |
| 29 | 3rtx_A | mRNA-capping enzyme | 7.5E-13 | 202 | 45–264 | 25–257 |
| 31 | 1p16_A | GTP–RNA, mRNA capping | 2.3E-13 | 205 | 43–259 | 19–259 |
| 32 | 1a0i_A | DNA ligase; ATP-dependent | 6.5E-12 | 152 | 88–246 | 26–242 |
| 33 | 3kyh_C | mRNA-capping enzyme | 2.3E-12 | 204 | 44–259 | 23–267 |
| 34 | 4xrp_B | RNL; RNA repair, kinase | 1.2E-06 | 199 | 43–244 | 5–316 |
The level of sequence similarity of Pab1020 to the different proteins carrying a NTase domain was revealed using HHpred implemented at www.toolkit.tuebingen.mpg.de. Homologies were detected using a highly sensitive HMM-HMM comparison mode with default settings. Searched database used was “pdb70_12jun16. Similar results were obtained using Pfam and COG databases.
Differently from monomeric DNA ligases, Pab1020 (Rnal3) ligases form a homodimer, which is a very rare feature among polynucleotide ligases and warrants further attention. Several features predict that this dimer interface is critical for Pab1020 complex assembly and/or function. For instance, PDBePISA analysis showed that the interface formed between 2 monomers has an interface area of ∼2274 Å2, with a high complex formation significance score of 0.518. Up to 58 residues (15.5% of total) interact between the 2 Pab1020 monomers. Interestingly, these residues are not randomly located within the Pab1020 polypeptide but, on the level of the primary sequence, are mainly found on either side of the catalytic domain.22,24 One particularly interesting interface residue is Gly296 that is strictly conserved among the Pab1020 family members (several additional residues of the domain interface are also evolutionary conserved). This residue is part of a C-terminal dimerization domain composed of 3 α-helices found at the dimer interface and generates a kink in the helix α10. Our dynamic light scattering studies indicated a hydrodynamic radius of approximately 10 nm for the Pab1020 wild type protein while the mutants G296A and ΔC-ter formed higher molecular weight molecules without precipitating. Therefore we suppose that the dimerization and C-terminal domains of Pab1020 are required for optimal assembly of the active homodimer. A similar suggestion has very recently been made for the M. thermoautotrophicum RNA ligase.25 Note also that the C-terminal domain of Pab1020 forms additional contacts with the other monomer, particularly in the proximity of the active site.
Polynucleotide ligase, but not nucleic acid binding, activity of Pab1020 is specific for RNA
The nucleic acid binding activity of Pab1020 was investigated by electrophoretic mobility shift (EMSA) assays using various Cy5-labeled nucleic acid substrates. Binding reactions were performed using 5′-dephosphorylated substrates to prevent any substrate circularization and/or ligation activities during EMSA assays (Fig. 1A). Our results revealed that Pab1020 formed a well-defined oligonucleotide-ligase complex with ssRNA oligonucleotides under stoichiometric conditions. Under these experimental conditions, some ssDNA binding was also observed (Fig. 1A). We then investigated the ligase activity of Pab1020 at 50°C after 5′-phosphorylating oligos used for binding assays, including Mn2+ as divalent cation in a standard activity buffer. Our results revealed that Pab1020 only showed circularization activity on ssRNA, while no activity was observed with ssDNA (or the different RNA/DNA homo- or hetero- oligonucleotide duplexes tested) used for binding assays (Fig. 1B). In these experiments, the circular RNA product migrated “faster” than the linear substrate ssRNA, in agreement with earlier studies.25 In these assays, the “catalytic” K95G mutant of RNA ligase Pab1020 was inactive, thus confirming that RNA specific circularization was indeed catalyzed by Pab1020. The Pab1020 G296A variant and the NTase domain alone still had weak RNA binding activity (Fig. 1C). We also stress that Pab1020 mutants G296A and “NTase domain” were inactive in circularization assays, albeit they still possessed a very weak adenylating activity, as witnessed by a very faint band migrating “slower” than the linear RNA substrate (Fig. 1D). These results suggest importance of the dimerization and/or the C-terminal domains for RNA circularization activity.
Figure 1.
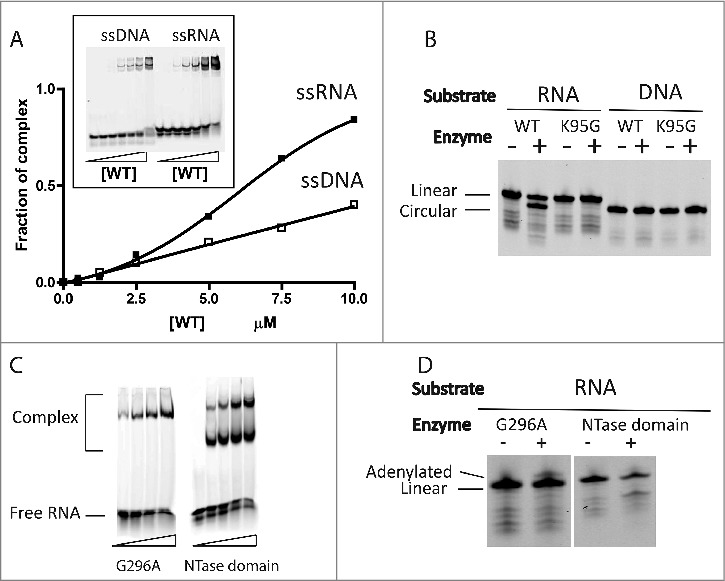
Pab1020 RNA ligase binds single-stranded DNA and RNA, but only circularizes single-stranded RNA oligonucleotides. (A) EMSA assays were performed with internally labeled (Cy5) single stranded DNA or RNA oligonucleotides using increasing amounts of wild-type (WT) Pab1020 RNA ligase. The relative amount of bound DNA or RNA was plotted against the protein concentration. Insert: On the EMSA gel, the amount of the higher molecular weight bands, corresponding to Pab1020-nucleic acid complexes, increased as a function of the protein concentration. (B) RNA and DNA ligation assays with WT and mutant K95G of Pab1020 RNA ligase. Standard ligation reactions containing 10 pmol Cy5-RNA or -DNA molecules and 200 pmol RNA ligase Pab1020 were incubated 90 min at 50°C. Reaction products were resolved on denaturing PAGE and a 700 nm scan of the gel was performed on Licor Odyssey Infrared Imager. While no activity was observed with DNA substrate, Pab1020 RNA ligase circularized an RNA oligoribonucleotide as shown on the gel with the apparition of a lower band corresponding to circular RNA molecules. Expectedly, a control reaction with an inactive enzyme (mutant K95G) presented no lower band. (C) Identical to panel (A), except that the enzymes used in the EMSA assays corresponded to the mutant G296A (dimerization domain) and the amino-terminal domain of 250 residues carrying a nucleotide transferase (NTase) domain. Both mutants were able to form RNA-Protein complexes with 18-mers single-stranded RNA. (D) Identical to panel (B), except that circularization was performed only with RNA substrate and with G296A mutant and NTase domain. No circRNAs were observed (positive control is indicated in panel B).
Pab1020 interacts with circular RNAs in cell-free extracts
As our biochemical studies confirmed that Pab1020 indeed acts as RNA ligase, we further investigated the substrate specificity of this protein in the cell. For these studies, 2 different RNA samples were analyzed using an experimental and computational RNA sequencing pipeline (Fig. 2A). For these studies a total RNA sample was extracted from a stationary phase culture of P. abyssi while a second sample was obtained by co-immunoprecipitation of RNA ligase Pab1020 after formaldehyde crosslinking between Pab1020 and cellular RNAs (RIP assay). Note that the affinity purified polyclonal antibody used for pull-down experiments revealed a single band in Western immunoblots of P. abyssi cell-free extracts (Fig. S1B). Isolated RNase III-fragmented RNA was used to prepare a RNA-seq library and sequenced following the Ion Torrent PGM RNA-seq protocol. Typical sequencing runs yielded ∼400 000 reads with a read size of 80 to 90 base pairs. All these reads were mapped to the P. abyssi reference genome reference (NC_000868, 1765118 base pairs) using Blastn. The unique criterion of the inversion of 2 matches within the same locus (Fig. 2A) led to the identification of approximately 80 000 putative circular RNAs suggesting up to 30 000 distinct junctions. To enrich for circRNAs, we used RNase R that specifically degrades linear RNA molecules in a 5′-3′ direction. For the total RNA fraction, our data revealed approximately 285 000 reads that resisted RNase R treatment and were mapped to the genome. Note that we still obtained linear reads using RNase R treated samples indicating that RNase R did not degrade all linear RNA molecules under these conditions (Table 2). However, 11 to 15 % of reads obtained using an RNase R enrichment were classified as “circular” using our computational criteria (see material and methods). These circular reads covered only a minor part of the transcribed genome (Table 2), therefore indicating that the combined experimental and computational criteria are strict.
Figure 2.
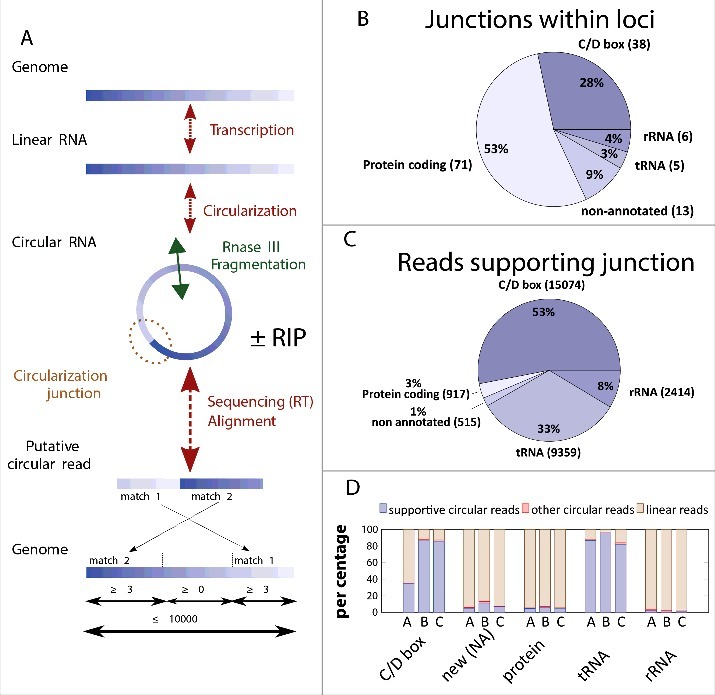
Identification of P. abyssi circRNAs using high throughput sequencing. (A) The workflow for identification of circularization junctions using RNA samples isolated from P. abyssi cells using IonTorrent semiconductor-based sequencing technology is shown. “ ± RIP” refers to the fact that identical computational approach was used for total and RNA immunoprecipitation (RIP) samples. Obtained linear and circular RNA molecules were fragmented at least once (indicated by a double arrow in panel A) using RNase III treatment. Following reverse transcription, samples were sequenced and obtained reads were aligned to the P. abyssi reference genome using Blastn. Reads were considered circular if 2 permuted matches covering the whole read was detected. (B) Number and percentage of the different functional classes (loci) considered circular in our sequencing experiments. (C) Number and percentage of the sequencing reads (total 28 279) supporting circularization of the different functional groups. (D) Percentage of the reads supporting RNA circularization (supportive circular reads) of the different RNA categories. Only intron containing tRNAs as identified as circular were included in the analysis. “Other circular reads” refers to a minority of putative circular reads that fulfill all the computational criteria indicated in panel A without supporting the junctions identified in panel B. Samples used were: A, circular reads after RIP assays using Pab1020 antibodies; B, circular reads after RIP assay and ribonuclease R treatment; C, circular reads in total RNA samples treated with ribonuclease R. New (NA) refers to previously non-annotated loci.
Table 2.
The summary of the RNA-seq results and RIP assays using the Pab1020 antibody.
| Circular reads |
Linear reads |
|||||
|---|---|---|---|---|---|---|
| Abbreviation | Sample | % of mapped reads | % of genomea | % of mapped reads | % of genome | Number of mapped reads |
| A | RIP | 8.3 | 6.8 | 91.7 | 50.3 | 247060 |
| B | RIP + RNase R | 14.7 | 2.7 | 85.3 | 8.7 | 44413 |
| C | Total RNA + RNase R | 11.4 | 2.8 | 88.6 | 7.3 | 284856 |
We are aware that our experimental and data analyses protocols may be prone for unwanted artifacts. Hence, to establish more selective criteria for circRNA identification we first merged the sequencing data from all of our experiments to identify the maximum number of circular RNAs. We observed that the circularization junctions were frequently shifted by 1 to 3 nucleotides between the different reads. This may either reflect the slight heterogeneity in choosing the transcription initiation site, or, alternatively, the presence of an identical base in 5′ and 3′ termini of a transcript that cannot be solved during the read mapping. Thus, we grouped together those putative circular reads where the circular junctions were located within 3 nucleotides. To be classified as circular, we only selected junctions that were identified at least in 2 independent experiments and supported each time by more than 3 individual reads that may have different start and end positions. We also requested that at least half of these putative circular reads that aligned entirely inside a given putative junction supported the junction.
With these strict and multiple constraints, we identified in P. abyssi a total of 133 individual circRNA loci (Fig. 2B) supported by 28 279 circular reads (Fig. 2C). The large majority of these RNA molecules interacted with Pab1020 in pull down experiments (Table 2, see also discussion).
Identification of functional categories of circular RNAs
The 133 P. abyssi circRNA loci represent 5 distinct functional groups: C/D Box RNA, non-annotated small RNAs, protein coding RNA, tRNA and rRNA (Fig. 2B). Among these, circular reads were overrepresented for 38 circular C/D Box and 5 tRNA molecules. Although 71 loci out of 133 loci corresponded to protein coding mRNAs, these were supported only by 3% of the analyzed reads (Fig. 2C). This approach also led to the discovery of 13 new circular RNAs was also identified [marked as non-annotated (NA) in Fig. 2B and C].
As pointed out by Danan et al,10 it is necessary to be attentive in attributing positions of circular junctions that are possibly influenced by reverse transcription, sequencing and mapping errors. The constraints applied in our computational pipeline (see materials and methods), allow attaining highly selective circRNAs identification. Numerous circRNAs, including non-coding RNAs, i.e. C/D box RNA, tRNA-intron and rRNA, were also observed in previous study.10
We also noticed that circRNAs corresponding to the different functional categories did not behave identically in RNase R-enrichment experiments. Strikingly, the relative portion of circular reads markedly increased after RNase R treatment from 35% to 86% for C/D Box RNAs and was constantly high, around 88%, for tRNAs (Fig. 2D). The fact that RNase R treatment induces an enrichment in the amount of reads supporting circularization junctions in pull-down and total RNA samples further indicates that Pab1020 RNA ligase specifically associates with circular RNA loci in P. abyssi cells. For the 3 additional functional groups, this RNase R enrichment for circRNAs was less obvious (Fig. 2D). Note that for the specific case of the tRNA-Trp, the circularization of the encoded-intron occurs simultaneously during the splicing process and linear intron intermediates is not expected to occur. For others RNAs (NA, protein coding and rRNA), the amount of circular reads is too low compared with linear reads for a same locus to allow the enrichment visualization. However, 3 non-annotated circRNAs (NA7, NA12, NA13 in Table 3) out of 13 showed some enrichment supported by significant amount of reads (Table 3). A high number (∼38 000) of circular reads were mapped to rRNAs (5S, 7S, 16S and 23S rRNAs) but in most cases, localization of the precise position of the junction point from permutated reads was far from evident, possibly reflecting the length and highly structured nature of these RNAs that hinders activity of the reverse transcriptase. However, in the case of the 5S rRNA, we identified 170 permuted reads indicating a specific circularization event between the 5′ and 3′ extremities of 5S rRNA (with a 10 nucleotide margin). As 5S rRNA interacted with Pab1020 in cell-free extracts and its circular form has been previously observed,10 this enzyme may participate in 5S rRNA pre-processing via a circular intermediate, as previously proposed for 16S and 23S rRNAs.15
Table 3.
List of 42 highly significant circular RNA molecules interacting with Pab1020 RNA ligase in cells. A summary of these results in the form of a Venn diagram is presented in Fig. 4.
| CircularRNA |
||||
|---|---|---|---|---|
| Locus | Start | End | Size | Enrichmenta |
| 38 C/D box RNAs | ||||
| sR46 | 57374 | 57441 | 67 | 1.05 |
| PabsnRNA44 (sR45) | 64244 | 64306 | 62 | 7.08 |
| PabsnRNA21 (sR14) | 65218 | 65278 | 60 | 2.87 |
| sR22 | 65333 | 65397 | 64 | 2.41 |
| sR32 | 67951 | 68012 | 61 | 1.97 |
| PabsnRNA3 (sR21) | 127067 | 127128 | 61 | 15.73 |
| PabsnRNA10 (sR2) | 230632 | 230696 | 64 | 1.94 |
| sR49 | 235439 | 235502 | 63 | 1.29 |
| PabsnRNA33 (sR13) | 245974 | 246035 | 61 | 3.86 |
| sR31 | 258066 | 258127 | 61 | 2.16 |
| PabsnRNA35 (sR29) | 318118 | 318182 | 64 | 2.81 |
| PabsnRNA32 (sR4) | 473175 | 473236 | 61 | 52.2 |
| PabsnRNA38 (sR58) | 541770 | 541831 | 61 | 2.15 |
| PabsnRNA40 (sR39) | 543855 | 543917 | 62 | 4.6 |
| PabsnRNA31 (sR20) | 553656 | 553721 | 65 | 2.67 |
| PabsnRNA12 (sR26) | 631518 | 631581 | 63 | 5.74 |
| PabsnRNA39 (sR60) | 631584 | 631644 | 60 | 7.49 |
| PabsnRNA13 (sR44) | 636702 | 636762 | 60 | 13.58 |
| PabsnRNA17 (sR7) | 648165 | 648228 | 63 | 43.0 |
| sR25 | 675408 | 675468 | 60 | 62.5 |
| PabsnRNA36 (sR55) | 910497 | 910569 | 72 | 4.91 |
| PabsnRNA27 (sR35) | 949199 | 949261 | 62 | 2.83 |
| sR56 | 960309 | 960370 | 61 | 5.31 |
| PabsnRNA25 (sR3) | 991446 | 991505 | 59 | 13.74 |
| PabsnRNA1 (sR24) | 1026074 | 1026133 | 59 | 6.93 |
| sR53 | 1042251 | 1042319 | 68 | 1.11 |
| PabsnRNA46 (sR38) | 1065728 | 1065791 | 63 | 5.14 |
| PabsnRNA28 (sR37) | 1195780 | 1195842 | 62 | 2.32 |
| PabsnRNA23 (sR1) | 1209270 | 1209329 | 59 | 36.27 |
| PabsnRNA34 (sR59) | 1260126 | 1260195 | 69 | 3.21 |
| sR41 | 1292356 | 1292415 | 59 | 5.63 |
| PabsnRNA5 (sR11) | 1397126 | 1397188 | 62 | 28.62 |
| PabsnRNA29 (sR8) | 1403662 | 1403723 | 61 | 113.04 |
| PabsnRNA42 (sR36) | 1409130 | 1409196 | 66 | 9.73 |
| PabsnRNA41 (sR48) | 1468649 | 1468712 | 63 | 1.23 |
| PabsnRNA6 (sR6) | 1536388 | 1536449 | 61 | 4.33 |
| PabsnRNA45 (sR34) | 1755871 | 1755930 | 59 | 3.02 |
| PabsnRNA9 (sR12) | 1755929 | 1755991 | 62 | 6.88 |
| 1 tRNA (intron) | ||||
| Pabt35 | 1330513 | 1330584 | 71 | 0.89 |
| 3 non-annotated (NA) new circular RNAs | ||||
| NA7 | 622461 | 622526 | 65 | 8.39 |
| NA12 | 1011096 | 1011158 | 62 | 1.91 |
| NA13 | 1674520 | 1674581 | 61 | 5.36 |
An enrichment factor for circular molecules was estimated using the ratios of the circular and linear reads in the pulldown fraction (PA) and the total RNA (PC) samples before and after RNase R treatment. The values shown were obtained by dividing PA by PC. Values higher than 1 indicate the enrichment for circRNAs due to selective degradation of linear RNA molecules.
Circularization of physiologically significant RNA molecules by an archaeal RNA ligase Pab 1020
To test whether RNAs interacting with Pab1020 in the cells may correspond to physiologically significant substrates of this enzyme, we assayed the ligation activity of Pab1020 RNA ligase using the linear fluorescent Cy5-RNA transcripts for 3 circRNAs identified during this work (Fig. 3). For these biochemical studies, we choose 2 Box C/D RNAs (SR4 and SR29) and the 5S rRNA as RNA-seq indicated that their circular isoforms exist in the cells and they specifically interacted with Pab1020 RNA ligase in pull-down experiments. Fluorescent RNA substrates were prepared by in vitro transcription with T7 RNA polymerase capable of incorporating Cy5-labeled nucleotide analogs.
Figure 3.
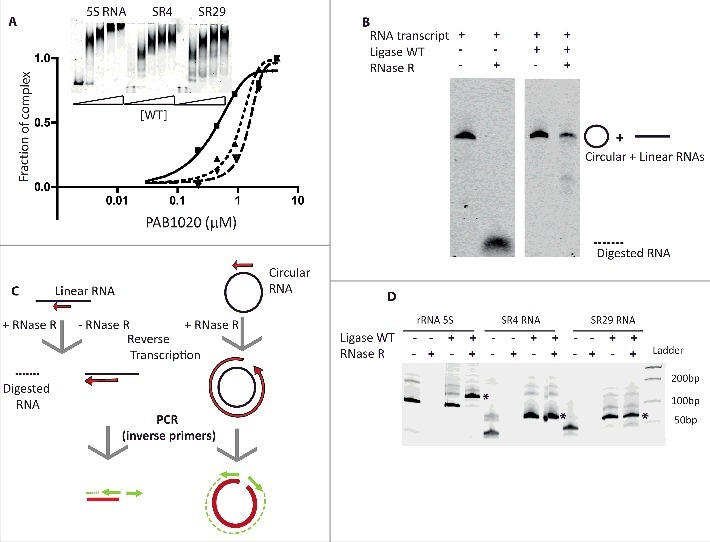
Pab1020 RNA ligase circularizes physiologically relevant RNA molecules. (A) RNA binding between Pab1020 RNA ligase (0.2 to 4.5 μM) and the in vitro transcripts (0.4 μM) corresponding to BoxC/D RNAs SR4 (▪) and SR29 (▴) and 5S rRNA (▾) was analyzed by EMSA. A fraction of protein-RNA complex formed was plotted as a function of input protein. Insert: On the EMSA gel, the amount of the higher molecular weight bands, corresponding to Pab1020-nucleic acid complexes, increased as a function of the protein concentration. (B) In vitro transcript of 5S rRNA was incubated (right panel) or not (left panel) with Pab1020 RNA ligase (WT) for 120 min at 55°C. After incubation, recovered RNAs were treated or not with exoribonuclease RNase R for 120 min at 37°C before analysis on a 7% acrylamide 8M urea gel. (C) Schematic illustration of RT-PCR experiments on linear and circular RNAs with divergent primers to distinguish linear RNAs from circular RNAs products after incubation with Pab1020 RNA ligase. Only reverse transcription and PCR reactions on a circular RNA template will lead to the total amplification of the substrate sequence. (D) cDNA generated using outward facing primers on RNAs previously incubated (+) or not (−) with Pab1020 RNA ligase and in the presence (+) or absence (−) of RNase R were separated by gel electrophoresis. A full-length product attesting to amplification of circular RNA molecules, indicated by the asterisk, was observed for 5S rRNA (128 bp), Box C/D SR4 RNA (68 bp) and Box C/D SR29 RNA (66 bp). Circularization was observed only in the presence of Pab1020 RNA ligase.
EMSA assays indicated that the 3 analyzed transcripts [Box C/D RNAs SR4 and SR29 (69mers) and the 5S rRNA (122mers)] formed specific RNA-protein complexes at near stoichiometric conditions (Fig. 3A). We also tested whether Pab1020 RNA ligase catalyzed the intramolecular ligation of the aforementioned transcripts (circularization) at 50°C. Fig. 3B shows that 15% acrylamide denaturing gel was not able to resolve the substrates and products of the RNA circularization reactions for these longer RNAs, as we successfully demonstrated in Fig. 1 for synthetic oligoribonucleotides. Therefore, to further identify circular RNA molecules, we used RNase R exoribonuclease treatment to discriminate between circular products and linear substrate RNAs. We observed that fluorescent transcript corresponding to P. abyssi 5S rRNA became partially resistant to RNase R treatment after incubation with Pab1020, whereas the linear substrate RNA was totally degraded (Fig. 3B). We further confirmed the RNA ligation activity of Pab1020 on the 5S rRNA and Box C/D RNAs SR4 or SR29 using inverse PCR. Briefly, the RNA ligation reactions were treated with RNase R, followed by the reverse transcription of each RNA (Fig. 3C). The outward facing (inverse) primers (when compared with the genomic sequence) were expected to amplify only circular templates, whereas only a small RT-product would be observed on a linear RNA template (Fig. 3C). For the same 3 selected RNAs (Box C/D RNAs SR4 orSR29 and 5S rRNA), after incubation with Pab1020 RNA ligase, we performed RT-PCR with the divergent primers described above. Indeed, we observed a full-length RT-PCR product (indicated by the asterisk in Fig. 3D) confirming RNA circularization by Pab1020. As negative control, in absence of RNA incubation with Pab1020 RNA ligase, the full-length amplification products corresponding to circular molecules (5S, SR4, SR29) were not observed (Fig. 3D).
These results from RNase R treatments and inverse PCR amplifications, confirmed that the RNA ligase encoded by Pab1020 gene is a (hyper)thermophilic protein that catalyzes the intramolecular ligation of RNA molecules.
Discussion
In archaea, 2 different major families of RNA ligases have been described. RtcB has been mainly implicated in ligation of single stranded tRNA halves with 2′-3′-cyclic phosphate and 5′-OH that occurs during splicing of pre-tRNAs. The Rnl3 ligase family, represented by Pab1020 studied here, uses similar mechanism as DNA ligases where 3′-OH reacts with 5′-phosphate to circularize RNA molecules. As the abundant presence of circular RNAs in all kingdoms of life, including hyperthermophilic Archaea, has only recently emerged, we investigated physiologic significance of Pab1020 RNA ligase activity previously observed only using synthetic substrates. The specific goal of our studies was to identify bona fide substrates of the Rnl3 family of RNA ligases. Using EMSA assays, we observed not only binding of Pab1020 to ssRNA (Fig. 1A), but to ssDNA as well. Nevertheless, under these experimental conditions, circularization activity was specific for oligoribonucleotides (Fig. 1B). Experiments shown in Fig. 1C also agree with the previous observations suggesting that “dimerization” and “C-terminal” domains of the Rnl3 family members are critical for intramolecular RNA circularization activity.25
We next established an experimental and computational pipeline to identify linear (substrates) and circular (products) RNA molecules that specifically interact with Pab1020 in cells. Toward this goal, we first isolated total RNA or Pab1020 interacting RNA molecules from P. abyssi cells. Obtained RNA samples were then reverse transcribed and sequenced using Ion Torrent technology. During this experimental protocol, RNase III fragmentation of circular molecules was necessary to allow ligation of adapters required for high-throughput sequencing. In the cases were this fragmentation occurs close to the junction and/or the reverse transcription does not proceed to the junction, reads originating from circular molecules would be erroneously classified as linear reads. However, although we are likely to underestimate the number of “circular” reads, the precise fragmentation site differs for individual molecules and it remains unlikely that all circular reads for a given locus would be missed in our computational analysis. Please note that our computational pipeline is ideally suited for analyses on prokaryotic data sets where RNA splicing is rare, as the frequent splicing would introduce gaps inside the matches.
The obtained sequencing reads were analyzed using the computational criteria described in Fig. 2A to identify the inverted matches, indicative of RNA circularization. At the first stage of our analysis, our experiments cumulatively supported a total of 30 000 distinct putative circularization junctions. For a given locus, the ratio between circular and linear reads varied substantially from approximately 5% up to 91%, indicating large in vivo heterogeneity in efficiency of RNA circularization process. This observation suggests that RNA circularization may be regulated and/or favored in cases where 5′ and 3′ extremities are brought together e.g., by structural constraints. The highest amount of circular reads (91% of all reads) was found for the intron of tRNA-Trp that carries the C/D box motif. In this case, 5′ and 3′ extremities are maintained in a close proximity by a bulge-helix-bulge structure already before ligation, thus likely favoring intramolecular ligation.
The Venn diagram shown in Fig. 4 illustrates the combined results from 3 individual experimental conditions of RNA-seq experiments that altogether revealed 133 circular loci (black numbers in Fig. 4). Interestingly, 127 of these circular loci (95%) were found in an RNA ligase pull-down fraction, suggesting that Pab1020 is necessary for RNA circularization in P. abyssi cells. We also stress that both linear and circular RNA molecules co-precipitated with the Pab1020 RNA ligase (Table 2). Note that the intersection of the RIP assays (A), RIP assays with RNase R treatment (B) and total RNA with RNase R treatment degrading linear molecules (C) contained approximately 40% of all the circular RNA loci. In agreement with previous studies,12,16 the most common circular RNAs correspond to the Box C/D RNA (guiding rRNA methylation) that showed an enrichment in RNase R experiments (indicated with white letters in Fig. 4, see also Table 3 for complete listing). These loci also suggested the presence of 3 novel non-coding RNAs that are evolutionary conserved within Thermococcales, indicating their functional importance. rRNAs, including P. abyssi 5S rRNA, and some coding RNAs did not show obvious enrichment in RNase R experiments, but have been observed in a circular form also in previous studies [Table S1.10,30]. Formation of the circular 5S rRNA could be compatible with the proposed processing mechanism of the archaeal pre-5S-RNA possibly31 but it is unclear whether this potential circular form corresponds to an additional processing intermediate or the mature 5S rRNA.15 When linear isoforms of the naturally occurring C/D box and 5 S rRNAs were used in binding (Fig. 3A) and circularization (Fig. 3B and C) assays, we observed an activity that was an order of magnitude higher than was observed for simple, likely non-structured, oligoribonucleotides (Fig. 1). As we have not observed intermolecular ligation in our assays (Figs. 1 and 3), we conclude that Pab1020 is both necessary and sufficient for RNA circularization in vitro and in vivo. This notion is further supported by our observations indicating absence of tRNA splicing products in our pull-down reactions. Thus, 2 RNA ligase families are not interchangeable.
Figure 4.
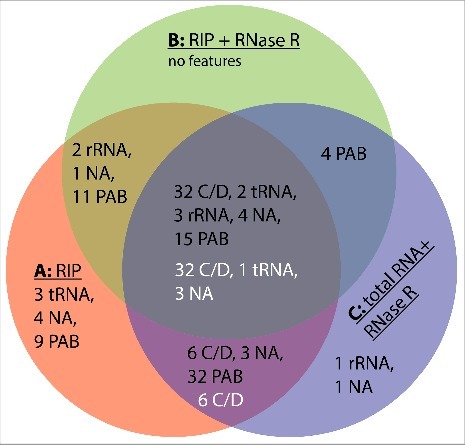
Venn diagram summarizing the results of our RNA-seq experiments. Samples used were: A, circular reads after RIP assays using Pab1020 antibodies; B, circular reads after RIP assay and ribonuclease R treatment; C, circular reads in total RNA samples treated with ribonuclease R. Black numbers refer to the categories of 133 junctions (Fig. 2B) and white numbers to 42 junctions with increased enrichment in RNase R experiments (Table 3).
We postulate that RNA circularization could provide increased thermostability by limiting thermal denaturation of stem structures formed between the 5′ and 3′ termini of box C/D RNAs, which is in agreement with the preferential presence of Pab1020 orhtologs in many extremophiles. However, we stress that thermal protection of small non-coding RNAs does not alone explain the functional importance of RNA circularization, as e.g., circular pseudouridylation guides carrying H/ACA motifs have not been reported to exist in archaea. The so called “H and ACA motifs” of these guides are obligatory found in single-stranded extremities of pseudouridylation guides,32,33 thus likely disfavoring intramolecular ligation.
In conclusion, here we have presented the combined results from pull-down experiments, RNA-seq experiments and in vitro circularization assays revealing that Pab1020 is the key enzyme required for RNA circularization in Archaea. Our results suggest the duplication and functional speciation of an ancestral NTase domain and/or DNA ligase toward RNA ligase activity and prompt for further characterization of the widespread functional roles of circular RNAs in prokaryotes.
Materials and methods
Strains and cell culture techniques
P. abyssi GE5 was grown in continuous culture in a gas lift bioreactor as described previously.34 Cells were collected in the exponential growth phase, followed by centrifugation at 6000 g for 15 min at 4°C. Strict anaerobic conditions were maintained during cell collection, centrifugation and storage of P. abyssi cells before further studies. Cell pellets were stored at −20°C.
Total RNA extraction from P. abyssi cells
Total RNA was isolated from approximately 108 P. abyssi cells following a single-step total RNA isolation protocol using the Tri-Reagent (Sigma-Aldrich). To remove contaminating DNA, 50 μg of isolated RNA was incubated with 50 units of RNase-free DNase I (New England Biolabs) for 30 min at 37°C. DNase I was inactivated by addition of 8 mM EDTA, pH 8 and 10 min incubation at 65°C. To obtain highly pure RNA samples, Tri-Reagent treatment was repeated to yield 40 μg of final RNA.
Production and affinity-purification of anti-Pab1020 antibodies
RNA ligase Pab1020 was produced and purified as described previously,22 except that the Cobalt-Hi-Trap column was replaced by a nickel column. Different Pab1020 mutants constructed during this work have been detailed in supplementary Materials and Methods S1.
6 mg of purified Pab1020 RNA ligase were used to immunize 2 New Zealand rabbits (Genecust, Luxembourg). We used 5 mL Hi-Trap N-hydroxy-succinimide (NHS)-activated column (GE Healthcare) for the affinity purification of the antibodies following the procedure described in the GE Healthcare Antibody Purification Handbook (http://www.gelifesciences.com/handbooks). Briefly, the Pab1020 protein was linked to the active groups of the column and 6 mL antisera from immunized rabbits were passed through the column. Proteins bound non-specifically to the column were eliminated using several washing steps. Fractions containing Pab1020 specific antibodies were collected using acid elution, immediately neutralized, dialyzed against phosphate-buffered saline and concentrated to 1.5 mL. Specificity and titer of the obtained antibodies were confirmed by Western Immunoblots (Fig. S1B).
Formaldehyde cross-link and RNA immunoprecipitation (pull-down) assays
Approximately 1010 P. abyssi GE5 cells were suspended in 25 mM HEPES pH 7, 15 mM MgCl2, 300 mM NaCl and were fixed using 2% formaldehyde during 20 min with gentle agitation. Crosslinking reactions were quenched using 100 mM glycine, followed by 2 successive washing steps in the same buffer as above. Obtained cell pellets were suspended in the extraction buffer containing 25 mM HEPES pH 7, 15 mM MgCl2, 300 mM NaCl, 0.4 M Sorbitol and complete, EDTA-free Protease cocktail (Roche). To obtain a soluble fraction containing crosslinked RNA samples, at this stage precipitates were eliminated by a 10-minute centrifugation step at 14 000 g. The obtained soluble fractions contained approximately 30 ng/μl RNA (estimated using A260 values) and 0.1 mg.mL−1 protein determined using a Bradford protein assay.
For the RNA immunoprecipitation (RIP) assays, to reduce or eliminate non-specific binding, 300 μL of the above supernatant were incubated for 1 hour at 4°C with 20 μL Protein A-agarose (Sigma-Aldrich), followed by centrifugation for 2 min at 10 000 g at 4°C. The resulting supernatant was incubated at 4°C for 3 hours with 5 μL of purified rabbit Anti-Pab1020 antibodies before addition of 20 μL Protein A-agarose for an additional hour. The pellet was recovered after centrifugation for 2 min at 10 000 g. RNA-protein-complexes bound to the beads were washed 3 times with 25 mM HEPES pH 7, 15 mM MgCl2, 300 mM NaCl and reversal of cross-links was achieved by incubation in the same buffer at 65°C for 1 hour. To recover RNA that specifically associated with the Pab1020 RNA ligase, samples were extracted with phenol/chloroform to remove proteins. For all samples, the remaining RNA was recovered using ethanol precipitation and dissolved in 20 μL water at a concentration of ∼10 ng.μL−1.
RNase R digestion of RNA samples
To enrich for circular RNA molecules, 100 ng of obtained RNA samples were treated with a magnesium dependent 3′ to 5′ exoribonuclease RNase R (Epicentre) at 37°C for 1 hour in a reaction buffer containing 20 mM Tris–HCl (pH 8), 0.1 mM MgCl2 and 100 mM KCl. RNase R treatments were performed with a ratio of 1 unit of enzyme per 10 ng of RNA. Ethanol precipitation was performed to remove the enzymes and salts, followed by a second RNase R treatment. Exoribonuclease resistant circRNA molecules were extracted with phenol/chloroform, ethanol precipitated and suspended in water at ∼10 ng.μL−1.
Experimental circRNA-seq workflow
RNase R treated and non-treated RNA samples were sequenced using the Ion Total RNA-seq Kit V2 (Life Technology). Total and Pab1020 associated RNA samples were used for high throughput sequencing studies. Briefly, cDNA libraries were prepared for each sample containing 100 – 800 ng of RNA that was treated using RNase III that cleaves inter- or intramolecular regions of double-stranded RNA.35 This resulted into formation of RNA fragments that were approximately 100-150 bp after 3 min incubation at 37°C with RNase III. RNA adaptor sequences were “splint ligated” to resulting linear RNA fragments using partly degenerate directional adapters. The first cDNA strand was reverse transcribed with the Superscript III Enzyme Mix (Life Technology) and double-stranded cDNA was amplified using Platinium PCR SuperMix High Fidelity (Life Technology) using manufacture's recommendations. Obtained DNA samples were diluted to obtain a final concentration of 100 pM, and were attached to beads and amplified using emulsion PCR. This circRNA protocol resulted into the clonal amplification of each RNA fragment within the microdroplets (Ion Spheres). Beads containing amplified DNA were enriched to eliminate empty spheres. Resulting samples were loaded onto an Ion 314 Chip V2 and sequenced in an Ion Personal Genome Machine Sequencer (PGM™, Life Technology). Polyclonal sequences originating from microbeads containing more than one template molecule were filtered out during automatic data processing using the dedicated IonTorrent server.
A computational pipeline for detection of putative circular reads
Sequencing reads were mapped to the P. abyssi GE5 reference genome (GenBank: NC_000868.1). Read mapping was performed using Blastn (version 2.2.26+)36 with the Megablast option using the following default parameters: word size at least 11, gap opening penalty of 5, gap extending penalty of 2, mismatch penalty of 3 and match reward of 1. The default expectation value threshold of 10 was used, and the maximum number of outputs was limited to 250 alignments per query. The maximum number of allowed outputs was not limiting our analyses, as the highest observed number of alignments for any given query was 182. To detect putative circular reads in sequencing data, all reads having 2 matches (from the Blastn output) that together covered the whole read, were selected. We considered only the permuted matches (Fig. 2A), with no overlap on the reference genome that were located within a 10 000 nucleotide window on the genome sequence. When more than 2 nucleotides and less than 11 (our minimum word size parameter) were missing in a match to cover the read, we looked “naively” for the small complementary match. This data processing step resulted into 2 sequence alignment data files in bam format that corresponded to linear and putative circular reads, respectively.
Electrophoretic mobility shift assay (EMSA)
Internally labeled RNA and DNA oligonucleotides were used for EMSA assays. The oligonucleotides used were: AUUCCGAUAG(Cy5dT)GACUACA (RNA) and ATTCCGATAG(Cy5dT)GACTACA (see also Table S2). Where indicated, RNA or DNA oligonucleotides or in vitro transcripts (2.5 μM) were incubated with protein samples (0.5–10 μM) in gel shift buffer containing 10 mM Tris–HCl pH 7, 150 mM NaCl, 0.5 mM DTT, 2.5 mM MgCl2, 0.01 mM ATP, 8 units of RNase Inhibitor (Biolabs). Binding reactions were performed at 50°C for 30 min in a final volume of 20 μL, and were analyzed using native gel electrophoresis. The final concentration of loading buffer used was 2 mM Tris–HCl pH 7.5, 10% glycerol, 0.1 mM EDTA pH 8, 20 μg.mL−1 BSA, 0.1 mg.ml−1 Orange C. Samples were loaded onto 10% native acrylamide/bisacrylamide gel (19:1) and electrophoresed in TEG 1X buffer (40 mM Glycine, 0.5 mM EDTA pH 8, 250 mM Tris–HCl) at 100 V for 3 h at room temperature. Gels were visualized and analyzed using an Odyssey system (LI-COR) using the 700 nm channel.
Preparation of fluorescent transcripts using in vitro transcription
Fluorescent transcripts for box C/D RNAs SR4 and SR29 (Table S2) were transcribed in vitro using synthetic DNA oligonucleotides as DNA template (Eurogentec). DNA templates were double stranded in the region corresponding to the 17 nucleotides of the T7 promoter sequence (TAATACGACTCACTATA). These dsDNA templates were prepared by hybridizing oligonucleotides corresponding to 10 μM T7 Promoter (plus strand), 10 μM T7 Promoter-RNA gene (PabsnRNA32 or PabsnRNA35, minus strand) in T7 RNA polymerase buffer (2,5X), by 3 min at 80°C, followed by slow cooling to ambient temperature. For 5S rRNA transcript, transcription was performed using 1 μg of DraI linearized pUC57 plasmid encoding T7 RNA polymerase promoter and 5S rRNA gene Pabr05.
Standard 20 μL transcription reactions contained dsDNA template (10 μM for box C/D RNA template or 0.1 μM for pUC57–5S RNA gene Pabr05), 7.5 mM of each NTP, 1X commercial reaction buffer, 12 units of RNasine, 0.25 mM Cy5-UTP and 2 μL Enzyme mix (T7 RNA Polymerase Megascript kit Ambion, Life Technology). All transcription reactions were allowed to proceed for 1 night at 37°C, before addition of 1 μL DNase Turbo (Megascript kit) and 15 min incubation at 37°C. Transcripts were analyzed using a 15% denaturing polyacrylamide gel electrophoresis (PAGE) and visualized by UV-shadowing. This allowed excision and elution of RNA from the gel with Maxam-Gilbert solution (0.5 M Na-acetate, 10 mM Mg-acetate, 1 mM EDTA, 0.1% SDS). RNA was precipitated using ethanol and transcripts were dissolved in diethylpyrocarbonate (DEPC) treated water to yield a final amount of approximately 700 pmol of transcript.
RNA circularization assays using fluorescent RNA and/or DNA oligonucleotides
Standard activity assays were performed in a mixture containing 10 mM Tris–HCl pH 7, 150 mM NaCl, 0.5 mM DTT, 2.5 mM MgCl2, 8 units of RNase Inhibitor (Biolabs), 10 pmol Cy5-RNA or -DNA molecules and 200 pmol RNA ligase Pab1020 in a total volume of 20 μL. Reactions were initiated by adding enzymes and incubated for 90 min at 50°C. Proteins were extracted with phenol/chloroform and obtained RNA samples were suspended in 10 μL H2O and 5μL denaturing buffer (Orange C 1 mg per mL in formamide). Reactions substrates and products were resolved using denaturing 18% PAGE containing 8M urea in 0.5X TBE. RNA molecules were revealed and quantified using an Odyssey imaging system as above (LI-COR).
RNA ligase assays on fluorescent box C/D and 5S RNA transcripts
Standard RNA ligase assays were performed as described above using Cy5-labeled box C/D or 5S transcripts. After incubation at 50°C for 120 min, RNA molecules were recovered by ethanol precipitation and resuspended in 10 μL of RNase R buffer 1X and incubated for 120 min at 37°C with 10 units of RNase R exoribonuclease (Epicentre), followed by 60 min incubation at 37°C with 40 μg of Proteinase K. Proteins were extracted by Phenol/chloroform treatment and RNA were recovered by ethanol precipitation and resuspended in 10 μL of water. RNase R resistant RNA molecules were detected and quantified using a 7% polyacrylamide gel containing 8M urea in 0.5X TBE. Reverse transcription and PCR (RT-PCR) with outward facing primers was also used to confirm RNA circularization. In this case, reaction products recovered either from RNase R treated or non-treated RNA ligation assays were reverse transcribed using M-MLV RT (50 units, Promega) and primers complementary to a central portion of box C/D RNA gene. cDNA templates were PCR amplified using Taq DNA polymerase and 2 divergent (outward facing) primers to anneal at the ends of the cDNA sequences. We performed 30 cycles of PCR and PCR products were visualized after electrophoresis on a 15% – 8 M urea polyacrylamide gels under denaturing conditions. Bands were visualized by ethidium bromide staining.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgements
The authors thank Joëlle Kuhn for production of Pab1020 antibodies. We are grateful to Ghislaine Henneke and Didier Flament for P. abyssi cells and thank Claire Toffano-Nioche, Marc Graille and Herman Van Tilbeurgh for many stimulating discussions and suggestions during this work. We also acknowledge Ursula Liebl for critical reading of the manuscript.
Funding
This work was supported by the Agence Nationale de la Recherche grant RETIDYNA. Work in our laboratory is supported by E. Polytechnique, CNRS and INSERM. Funding for open access charge: INSERM.
References
- 1.Ebbesen KK, Kjems J, Hansen TB. Circular RNAs: Identification, biogenesis and function. Biochim Biophys Acta 2015; 1859:163-8; PMID:26171810; https://doi.org/ 10.1016/j.bbagrm.2015.07.007 [DOI] [PubMed] [Google Scholar]
- 2.Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA 2015; 20:1829-42; PMID:25404635; https://doi.org/25259915 10.1261/rna.047126.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vicens Q, Westhof E. Biogenesis of Circular RNAs. Cell 2014; 159:13-4; PMID:25259915; https://doi.org/ 10.1016/j.cell.2014.09.005 [DOI] [PubMed] [Google Scholar]
- 4.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature 2013; 495:384-8; PMID:23446346; https://doi.org/ 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 5.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al.. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013; 495:333-8; PMID:23446348; https://doi.org/ 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 6.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet 2013; 9:e1003777; PMID:24039610; https://doi.org/ 10.1371/journal.pgen.1003777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell 2013; 51:792-806; PMID:24035497; https://doi.org/ 10.1016/j.molcel.2013.08.017 [DOI] [PubMed] [Google Scholar]
- 8.Monat C, Cousineau B. Circularization pathway of a bacterial group II intron. Nucleic Acids Res 2016; 44:1845-53; PMID:26673697; http://doi.org/11511373 10.1093/nar/gkv1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murray HL, Mikheeva S, Coljee VW, Turczyk BM, Donahue WF, Bar-Shalom A, Jarrell KA. Excision of group II introns as circles. Mol Cell 2001; 8:201-11; PMID:11511373; https://doi.org/ 10.1016/S1097-2765(01)00300-8 [DOI] [PubMed] [Google Scholar]
- 10.Danan M, Schwartz S, Edelheit S, Sorek R. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res 2011; 40:3131-42; PMID:22140119; https://doi.org/ 10.1093/nar/gkr1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salgia SR, Singh SK, Gurha P, Gupta R. Two reactions of Haloferax volcanii RNA splicing enzymes: joining of exons and circularization of introns. RNA 2003; 9:319-30; PMID:12592006; https://doi.org/ 10.1261/rna.2118203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh SK, Gurha P, Tran EJ, Maxwell ES, Gupta R. Sequential 2′-O-methylation of archaeal pre-tRNATrp nucleotides is guided by the intron-encoded but trans-acting box C/D ribonucleoprotein of pre-tRNA. J Biol Chem 2004; 279:47661-71; PMID:15347671; https://doi.org/ 10.1074/jbc.M408868200 [DOI] [PubMed] [Google Scholar]
- 13.Dalgaard JZ, Garrett RA. Protein-coding introns from the 23S rRNA-encoding gene form stable circles in the hyperthermophilic archaeon Pyrobaculum organotrophum. Gene 1992; 121:103-10; PMID:1427083; https://doi.org/ 10.1016/0378-1119(92)90167-N [DOI] [PubMed] [Google Scholar]
- 14.Lykke-Andersen J, Garrett RA. Structural characteristics of the stable RNA introns of archaeal hyperthermophiles and their splicing junctions. J Mol Biol 1994; 243:846-55; PMID:7966305; https://doi.org/ 10.1006/jmbi.1994.1687 [DOI] [PubMed] [Google Scholar]
- 15.Tang TH, Rozhdestvensky TS, d'Orval BC, Bortolin ML, Huber H, Charpentier B, Branlant C, Bachellerie JP, Brosius J, Hüttenhofer A, et al.. RNomics in Archaea reveals a further link between splicing of archaeal introns and rRNA processing. Nucleic Acids Res 2002; 30:921-30; PMID:11842103; https://doi.org/ 10.1093/nar/30.4.921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Starostina NG, Marshburn S, Johnson LS, Eddy SR, Terns RM, Terns MP. Circular box C/D RNAs in Pyrococcus furiosus. Proc Natl Acad Sci U S A 2004; 101:14097-101; PMID:15375211; https://doi.org/ 10.1073/pnas.0403520101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watkins H, Bohnsack M. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA 2012; 3:397-414; PMID:22065625; https://doi.org/ 10.1002/wrna.117 [DOI] [PubMed] [Google Scholar]
- 18.Randau L. RNA processing in the minimal organism Nanoarchaeum equitans. Genome Biol 2012; 13:R63; PMID:22809431; https://doi.org/ 10.1186/gb-2012-13-7-r63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clouet d'Orval B, Bortolin ML, Gaspin C, Bachellerie JP. Box C/D RNA guides for the ribose methylation of archaeal tRNAs. The tRNATrp intron guides the formation of two ribose-methylated nucleosides in the mature tRNATrp. Nucleic Acids Res 2001; 29:4518-29; PMID:11713301; https://doi.org/ 10.1093/nar/29.22.4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lykke-Andersen J, Aagaard C, Semionenkov M, Garrett RA. Archaeal introns: splicing, intercellular mobility and evolution. Trends Biochem Sci 1997; 22:326-31; PMID:9301331; https://doi.org/ 10.1016/S0968-0004(97)01113-4 [DOI] [PubMed] [Google Scholar]
- 21.Englert M, Sheppard K, Aslanian A, Yates JR 3rd, Soll D. Archaeal 3′-phosphate RNA splicing ligase characterization identifies the missing component in tRNA maturation. Proc Natl Acad Sci U S A 2011; 108:1290-5; PMID:21209330; https://doi.org/ 10.1073/pnas.1018307108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brooks MA, Meslet-Cladiere L, Graille M, Kuhn J, Blondeau K, Myllykallio H, van Tilbeurgh H. The structure of an archaeal homodimeric ligase which has RNA circularization activity. Protein Sci 2008; 17:1336-45; PMID:18511537; https://doi.org/ 10.1110/ps.035493.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chambers CR, Patrick WM. Archaeal Nucleic Acid Ligases and Their Potential in Biotechnology. Archaea 2015; 2015:170571; PMID:26494982; https://doi.org/ 10.1155/2015/170571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu H, Yoshinari S, Ghosh R, Ignatochkina AV, Gollnick PD, Murakami KS, Ho CK. Structural and mutational analysis of archaeal ATP-dependent RNA ligase identifies amino acids required for RNA binding and catalysis. Nucleic Acids Res 2016; 44:2337-47; PMID:26896806; https://doi.org/ 10.1093/nar/gkw094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Torchia C, Takagi Y, Ho CK. Archaeal RNA ligase is a homodimeric protein that catalyzes intramolecular ligation of single-stranded RNA and DNA. Nucleic Acids Res 2008; 36:6218-27; PMID:18829718; https://doi.org/ 10.1093/nar/gkn602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhelkovsky AM, McReynolds LA. Simple and efficient synthesis of 5′ pre-adenylated DNA using thermostable RNA ligase. Nucleic Acids Res 2011; 39:e117; PMID:21724605; https://doi.org/ 10.1093/nar/gkr544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhelkovsky AM, McReynolds LA. Structure-function analysis of Methanobacterium thermoautotrophicum RNA ligase - engineering a thermostable ATP independent enzyme. BMC Mol Biol 2012; 13:24; PMID:22809063; https://doi.org/ 10.1186/1471-2199-13-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Unciuleac MC, Goldgur Y, Shuman S. Structure and two-metal mechanism of a eukaryal nick-sealing RNA ligase. Proc Natl Acad Sci U S A 2015; 112:13868-73; PMID:26512110; https://doi.org/ 10.1073/pnas.1516536112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, et al.. CDD: NCBI's conserved domain database. Nucleic Acids Res 2015; 43:D222-6; PMID:25414356; https://doi.org/ 10.1093/nar/gku1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doose G, Alexis M, Kirsch R, Findeiss S, Langenberger D, Machne R, Mörl M, Hoffmann S, Stadler PF. Mapping the RNA-Seq trash bin: unusual transcripts in prokaryotic transcriptome sequencing data. RNA Biol. 2013; 10:1204-10; PMID:23702463; https://doi.org/ 10.4161/rna.24972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holzle A, Fischer S, Heyer R, Schutz S, Zacharias M, Walther P, Allers T, Marchfelder A. Maturation of the 5S rRNA 5′ end is catalyzed in vitro by the endonuclease tRNase Z in the archaeon H. volcanii. RNA 2008; 14:928-37; PMID:18369184; https://doi.org/ 10.1261/rna.933208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balakin AG, Smith L, Fournier MJ. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell 1996; 86:823-34; PMID:8797828; https://doi.org/ 10.1016/S0092-8674(00)80156-7 [DOI] [PubMed] [Google Scholar]
- 33.Ganot P, Bortolin ML, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 1997; 89:799-809; PMID:9182768; https://doi.org/ 10.1016/S0092-8674(00)80263-9 [DOI] [PubMed] [Google Scholar]
- 34.Pluchon PF, Fouqueau T, Creze C, Laurent S, Briffotaux J, Hogrel G, Palud A, Henneke G, Godfroy A, Hausner W, et al.. An extended network of genomic maintenance in the archaeon Pyrococcus abyssi highlights unexpected associations between eucaryotic homologs. PLoS One 2013; 8:e79707; PMID:24244547; https://doi.org/ 10.1371/journal.pone.0079707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicholson AW. Ribonuclease III mechanisms of double-stranded RNA cleavage. Wiley Interdiscip Rev RNA 2014; 5:31-48; PMID:24124076; https://doi.org/ 10.1002/wrna.1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics 2009; 10:421; PMID:20003500; https://doi.org/ 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


