Figure 2.
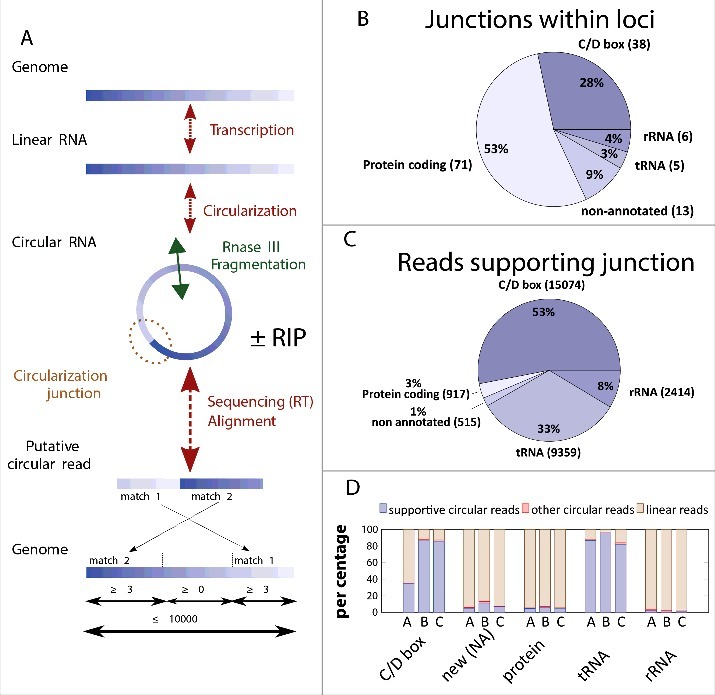
Identification of P. abyssi circRNAs using high throughput sequencing. (A) The workflow for identification of circularization junctions using RNA samples isolated from P. abyssi cells using IonTorrent semiconductor-based sequencing technology is shown. “ ± RIP” refers to the fact that identical computational approach was used for total and RNA immunoprecipitation (RIP) samples. Obtained linear and circular RNA molecules were fragmented at least once (indicated by a double arrow in panel A) using RNase III treatment. Following reverse transcription, samples were sequenced and obtained reads were aligned to the P. abyssi reference genome using Blastn. Reads were considered circular if 2 permuted matches covering the whole read was detected. (B) Number and percentage of the different functional classes (loci) considered circular in our sequencing experiments. (C) Number and percentage of the sequencing reads (total 28 279) supporting circularization of the different functional groups. (D) Percentage of the reads supporting RNA circularization (supportive circular reads) of the different RNA categories. Only intron containing tRNAs as identified as circular were included in the analysis. “Other circular reads” refers to a minority of putative circular reads that fulfill all the computational criteria indicated in panel A without supporting the junctions identified in panel B. Samples used were: A, circular reads after RIP assays using Pab1020 antibodies; B, circular reads after RIP assay and ribonuclease R treatment; C, circular reads in total RNA samples treated with ribonuclease R. New (NA) refers to previously non-annotated loci.
