Figure 1.
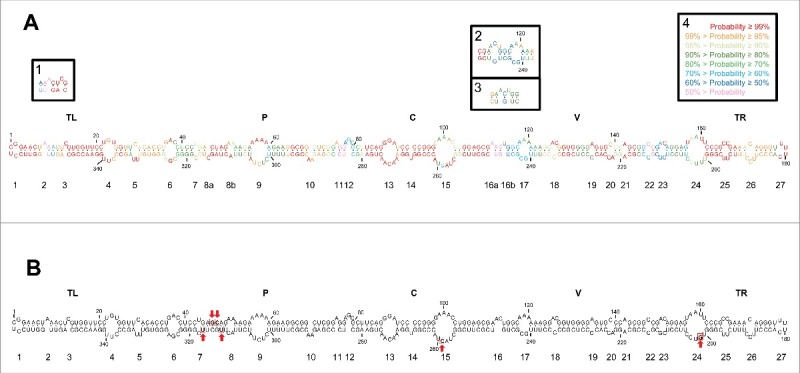
(A) Rod-like secondary structure of minimum free energy predicted by RNAstructure for the mc PSTVd (+) RNA of the NB variant. Insets (1) and (2), alternative motifs predicted by RNAfold and inset (3), alternative motif predicted by Mfold. Colors denote the probability (see inset 4) of a nucleotide being double- or single-stranded as predicted by RNAstructure. (B) Rod-like secondary structure of minimum free energy proposed for the mc PSTVd (+) RNA of the intermediate variant,82 which differs from NB in 6 substitutions: C46→G, U47→C, U201→G, U259→C, A313→U, and C317→U (within squares and marked with arrows). The structure of the intermediate variant, which is presented here for comparative purposes and as a reference for loop numbering,48 has been edited to remove an non-existing extra G-C base pair adjacent to the terminal left loop.48 Loops 8 and 16 in the intermediate variant are split in the NB variant into 8a and 8b, and 16a and 16b, respectively. TL (terminal left), P (pathogenicity), C (central), V (variable) and TR (terminal right) are structural domains defined previously.82
