Figure 7. Characterization of LIHC immune microenvironment using RNA-seq data.
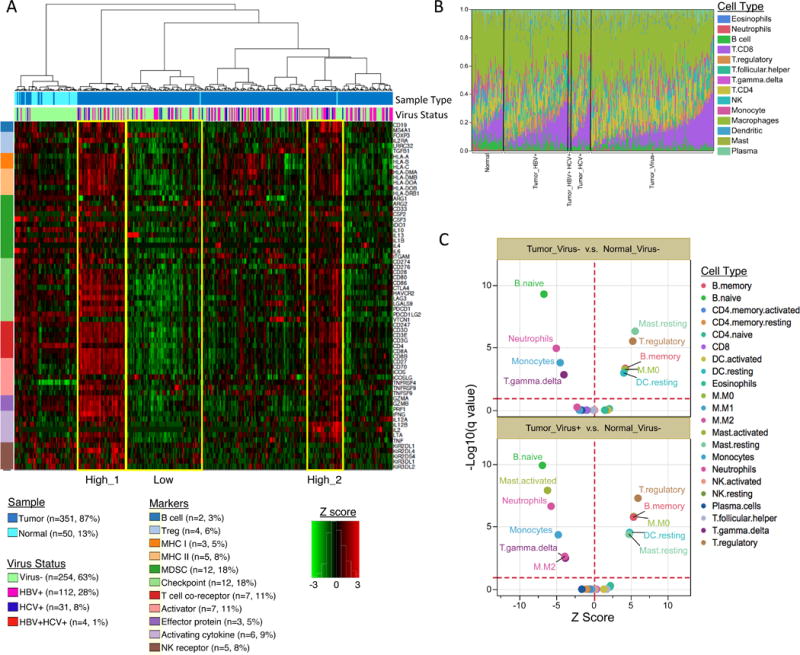
(A) Unsupervised hierarchical clustering of gene expression identifies immune profiles within HCC patients. Sixty-six manually curated immune cell markers were used for clustering. (B) The CIBERSORT-inferred relative fractions of different immune cell types varied across tumor and tumor adjacent normal samples and were not associated with virus status. (C) CIBERSORT cellular composition analysis revealed striking differences in relative compositions of immune cell populations between tumor and tumor-adjacent normal tissues. P values were calculated by Wilcoxon rank-sum test and adjusted for multiple testing (q value). The red dotted lines on the y axis indicate q value of 0.01. The red dotted lines on × axis indicates Z score of 0. The analysis was performed for all CIBERSORT immune cell types but only the significant ones are labeled on the plot.
