Fig. 2.
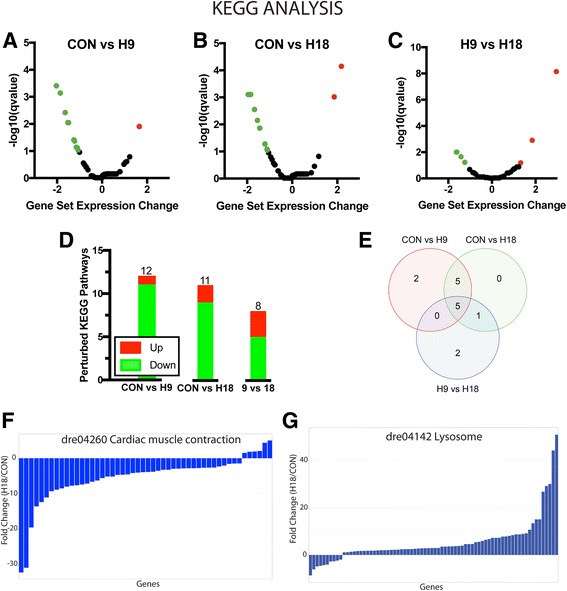
KEGG pathway classification of differentially expressed genes. Volcano plots of KEGG pathways in CON vs H9 (a), CON vs H18 (b), and H9 vs. H18 (c) comparisons. Gene set expression change (x-axis) of DEG within a KEGG pathway is a normalized enrichment score based on comparisons between different time points and relative to the entire DEG gene set. Significantly downregulated pathways are shown in green and significantly upregulated pathways are shown in red. d Distribution of significantly perturbed KEGG pathways in the three pair-wise comparisons. Significantly downregulated terms are shown in green. Significantly upregulated terms are shown in red. Cutoff for significance was q < 0.1 according to default values and parameters of the R package “gage” v2.22.0. e Venn diagram showing overlap of perturbed KEGG terms between CON, H9, and H18 sample comparisons. Cutoff for significance was q < 0.1 according to default values and parameters of the R package “gage” v2.22.0. Fold change (H18/CON) of the DEG of the dre04260 Cardiac muscle contraction (f) and dre04142 Lysosome (g) KEGG pathways
