Figure 1.
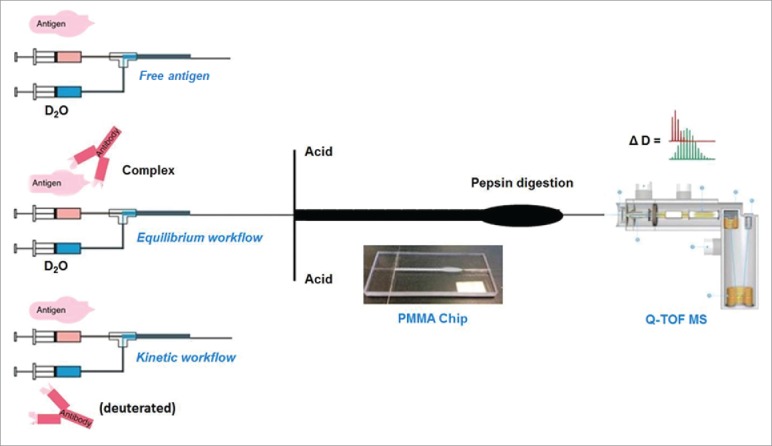
Schematic of the three TRESI-HDX experiments carried out in this work. All three workflows incorporate millisecond HDX labeling followed by acid quenching, digestion, on-chip electrospray ionization, MS detection and data analysis. (Top) TRESI-HDX of free antibody. Peptide-specific uptake data from this experiment are subtracted from‘equilibrium'or ‘kinetic’ experiment data to provide HDX difference profiles associated with complexation. (Middle) ‘Equilibrium’ workflow: Antibody and antigen are pre-equilibrated prior to analysis. (Bottom) ‘Kinetic’ workflow: Antibody is introduced to the antigen through the TRESI mixer, so that binding and labeling are initiated simultaneously.
