Figure 1. The synteny of antibiotic resistance genes provides historical context and foreshadows future.
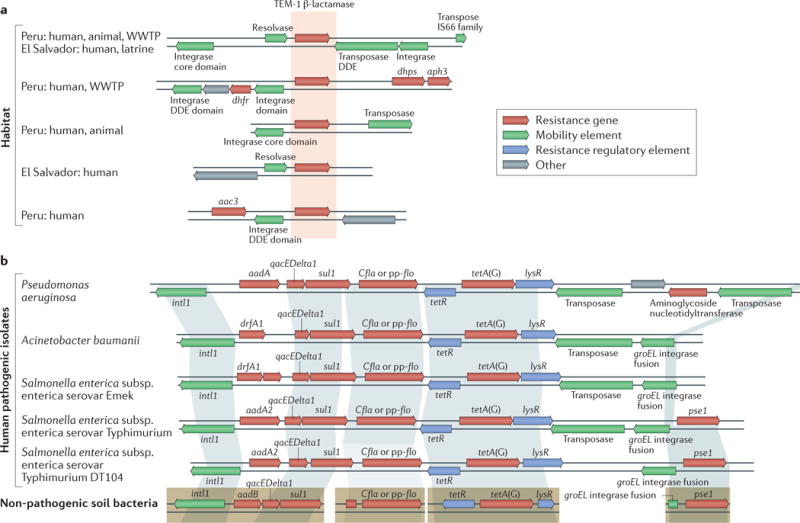
The genetic context of antibiotic resistance genes can provide important evidence regarding the likelihood of past or future horizontal gene transfer. a | A single β-lactamase (TEM-1) was found in 25 different genetic contexts in a recent cross-habitat resistome study, which provided a historical record of past mobility. Five contexts are shown here. Habitats in which a gene was discovered are indicated on the left (‘human’: human-associated microbial community; ‘animal’: animal-associated microbial community; ‘WWTP’: wastewater treatment plant; ‘latrine’: composting latrine). b | Four resistance determinants that were discovered in functional selections of soil metagenomic libraries (bottom) have high identity with resistance genes from clinical pathogenic isolates. Shading indicates >99% nucleotide identity, which suggests recent horizontal transfer. Resistance genes in pathogens are syntenic with mobility elements and other resistance genes, which suggests that they may be present on a mobile multidrug resistance cassette and are therefore likely to undergo horizontal gene transfer. DDE, Asp-Asp-Glu. Part a is adapted with permission from REF.66, Macmillan Publishers Limited. Part b is adapted with permission from REF.14, AAAS.
