Abstract
Phosphomannosylation is a modification of cell wall proteins that occurs in some species of yeast-like organisms, including the human pathogen Candida albicans. These modified mannans confer a negative charge to the wall, which is important for the interactions with phagocytic cells of the immune systems and cationic antimicrobial peptides. In Saccharomyces cerevisiae, the synthesis of phosphomannan relies on two enzymes, the phosphomannosyltransferase Ktr6 and its positive regulator Mnn4. However, in C. albicans, at least three phosphomannosyltransferases, Mnn4, Mnt3 and Mnt5, participate in the addition of phosphomannan. In addition to MNN4, C. albicans has a MNN4-like gene family composed of seven other homologous members that have no known function. Here, using the classical mini-Ura-blaster approach and the new gene knockout CRISPR-Cas9 system for gene disruption, we generated mutants lacking single and multiple genes of the MNN4 family; and demonstrate that, although Mnn4 has a major impact on the phosphomannan content, MNN42 was also required for full protein phosphomannosylation. The reintroduction of MNN41, MNN42, MNN46, or MNN47 in a genetic background lacking MNN4 partially restored the phenotype associated with the mnn4Δ null mutant, suggesting that there is partial redundancy of function between some family members and that the dominant effect of MNN4 over other genes could be due to its relative abundance within the cell. We observed that additional copies of alleles number of any of the other family members, with the exception of MNN46, restored the phosphomannan content in cells lacking both MNT3 and MNT5. We, therefore, suggest that phosphomannosylation is achieved by three groups of proteins: [i] enzymes solely activated by Mnn4, [ii] enzymes activated by the dual action of Mnn4 and any of the products of other MNN4-like genes, with exception of MNN46, and [iii] activation of Mnt3 and Mnt5 by Mnn4 and Mnn46. Therefore, although the MNN4-like genes have the potential to functionally redundant with Mnn4, they apparently do not play a major role in cell wall mannosylation under most in vitro growth conditions. In addition, our phenotypic analyses indicate that several members of this gene family influence the ability of macrophages to phagocytose C. albicans cells.
Keywords: cell wall, phosphomannosylation, Candida albicans, phagocytosis, phosphomannosyltransferase, CRISPR-Cas9 system, mini-Ura-blaster
Introduction
Infections caused by members of the Candida genus can result in both superficial and deep tissue invasion—the latter frequently being associated with immunocompromised patients resulting in high morbidity and mortality rates (Brown et al., 2012). Of the 190 species classified within the Candida genus, Candida albicans is the most frequent agent of candidiasis, although other Candida species are increasingly becoming a major cause of concern (Sanguinetti et al., 2015).
The C. albicans cell wall plays a key role in cellular fitness and as a molecular scaffold to which several virulence-related proteins are attached (Mora-Montes et al., 2009; Gow and Hube, 2012; Erwig and Gow, 2016). Indeed, mutant strains with defects in the synthesis of cell wall components often display virulence attenuation and alterations in immune recognition (Bates et al., 2005, 2006, 2013; Munro et al., 2005; Prill et al., 2005; Mora-Montes et al., 2007, 2010; Hall et al., 2013; Courjol et al., 2015). The Candida cell wall is composed of an inner core of chitin, β1, 3- and β1, 6-glucans, and an outer layer of highly glycosylated proteins that are rich in various classes of mannoligosaccharides (mannoproteins) (Klis et al., 2001; Hall and Gow, 2013). These glycoproteins have high molecular weight-branched mannose oligosaccharides linked to Asp (N-liked mannans) and shorter linear glycans attached to Ser/Thr (O-linked mannans) residues (Mora-Montes et al., 2009; Hall and Gow, 2013). In yeast-like organisms, such as Saccharomyces cerevisiae or C. albicans, both types of mannans can be modified by ether-linked mannosylphosphate (phosphomannan) (Mora-Montes et al., 2009; Orlean, 2012). The N-linked mannan has an oligosaccharide core synthesized in the endoplasmic reticulum, which is modified by Golgi-resident mannosyltransferases that elaborate the α1, 6-polymannose backbone, and lateral branched mannooligosaccharides that are α1, 2-, α1, 3- or β1, 2-linked (Martinez-Duncker et al., 2014). The O-linked mannans of up to seven units are composed predominantly of α1, 2-mannose sugars (Munro et al., 2005; Diaz-Jimenez et al., 2012). This phosphomannan moiety is believed to participate in the regulation of nucleotide transport across the Golgi membrane, in the cross-linking between cell wall proteins and glucan, and in stress regulation during stationary growth phase or under conditions of drought or high osmolality (Jigami and Odani, 1999). Charge neutral C. albicans mutant cells that lack cell wall phosphomannan showed increased resistance to the inhibitory effect of the cationic peptide DsS3(1-16) (Harris et al., 2009), and were less quickly phagocytosed by primary macrophages and macrophage cell lines (McKenzie et al., 2010; Lewis et al., 2012; Bain et al., 2014). These observations underscore the importance of this cell wall component during the host-fungus interaction.
Phosphomannan synthesis has been characterized in most detail in S. cerevisiae. The Golgi-resident Mnn6/Ktr6 protein is the sole phosphomannosyltransferase in this organism and responsible for the addition of phosphomannan to both N-linked and O-linked mannans (Wang et al., 1997). Although there has been no enzyme activity associated with Mnn4, this protein is presumed to be a positive regulator of the phosphomannosyltransferase, because overexpression or disruption of MNN4 positively and negatively affected the cell wall phosphomannan content, respectively (Odani et al., 1996; Jigami and Odani, 1999). Recently, Mnn14 has been also involved in the phosphomannosylation of S. cerevisiae N-linked mannans (Kim et al., 2017). This gene is a paralog of MNN4, and these proteins showed functional redundancy in addition of mannosylphosphate to the N-linked mannan core (Kim et al., 2017). In C. albicans, MNT3 and MNT5 encode for functional orthologs of S. cerevisiae MNN6/KTR6, with redundant phosphomannosyltransferase activity, and are involved in the elaboration of about 50% of the cell wall phosphomannan (Mora-Montes et al., 2010). This organism also contains a functional ortholog of S. cerevisiae MNN4, and the cell wall phosphomannan is barely detected in a C. albicans mnn4Δ null mutant (Hobson et al., 2004). In contrast to S. cerevisiae, the C. albicans phosphomannan moiety appears to function as a molecular scaffold for the addition of linear β1, 2-oligosaccharides of up to 14 mannose residues (Hobson et al., 2004; Mora-Montes et al., 2009). This participates in establishing the hydrophobic properties of the cell (Singleton et al., 2005). Interestingly, in C. albicans there is an extended MNN4-like gene family that is composed of seven additional MNN4-like genes of unknown function, named MNN41, MNN42, MNN43, MNN44, MNN45, MNN46, and MNN47 (Butler et al., 2009).
To investigate the function of the members of this gene family, we disrupted MNN4 along with the seven other orthologs and characterized their ability to bind Alcian Blue. We found that, aside from deletion of MNN4, only disruption of MNN42 was capable of reducing the gross cell wall phosphomannan content. However, overexpression of a range of MNN4-like genes in genetic backgrounds lacking either MNN4 or MNT3 and MNT5 showed that these genes could complement the mutant phenotype to some extent. These data suggest that all the MNN4-like genes family are capable of participating in the elaboration of C. albicans cell wall phosphomannan under permissive conditions.
Materials and methods
Strains and culturing conditions
Strains used in this work are listed in Table 1. Cells were growth in YPD medium (1% [w/v] yeast extract, 2% [w/v] gelatin peptone, 2% [w/v] dextrose) at 28°C and 200 rpm. When solid medium was required, 2% (w/v) agar was added. Yeast transformants were selected in SD medium (0.76% [w/v] yeast nitrogen base with ammonium sulfate without amino acids, 2% [w/v] dextrose and 0.077% [w/v] complete supplement mixture minus uracil] with 50 μg/mL uridine when required. For selection of transformants using the CRISPR-Cas9 strategy, cells were grown on YPD agar supplemented with 200 μg/mL nourseothricin (ClonNAT, WERNER BioAgents, Jena, Germany).
Table 1.
Strains used in this work.
| Strain | Genotype | References |
|---|---|---|
| CAI4 | ura3Δ::imm434/ura3Δ::imm434 | Fonzi and Irwin, 1993 |
| BWP17 | ura3Δ::imm434/ura3Δ::imm434, his1Δ::hisG/his1Δ::hisG, arg4Δ::hisG/arg4Δ::hisG | Wilson et al., 1999 |
| NGY152 | As CAI4, but RPSI/rps1Δ::Clp10 | Brand et al., 2004 |
| CDH7 | As CAI4, but mnn4Δ::hisG/mnn4Δ::hisG | Hobson et al., 2004 |
| CDH15 | As CDH7, but RPSI/rps1Δ::Clp10 | Hobson et al., 2004 |
| NGY522 | As CAI4, but mnt5Δ::higG/mnt5Δ::hisG; mnt3Δ::dp1200/mnt3Δ::dp1200 | Mora-Montes et al., 2010 |
| NGY1227 | As NGY522, but RPS1/rps1Δ::CIp10 | Mora-Montes et al., 2010 |
| HMY189 | As CAI4, but mnn41Δ::dp1200/mnn41Δ::dp1200, RPS1/rps1Δ:: Clp10 | This work |
| HMY81 | As CAI4, but mnn42Δ::dp1200/mnn42Δ::dp1200, RPS1/rps1Δ:: Clp10 | This work |
| HMY84 | As CAI4, but mnn42Δ::dp1200/mnn42Δ::dp1200, RPS1/rps1Δ:: Clp10-MNN42 | This work |
| HMY190 | As CAI4, but mnn43Δ::dp1200/mnn43Δ::dp1200, RPS1/rps1Δ:: Clp10 | This work |
| HMY191 | As CAI4, but mnn44Δ::dp1200/mnn44Δ::dp1200, RPS1/rps1Δ:: Clp10 | This work |
| HMY192 | As CAI4, but mnn45Δ::dp1200/mnn45Δ::dp1200, RPS1/rps1Δ:: Clp10 | This work |
| HMY193 | As CAI4, but mnn46Δ::dp1200/mnn46Δ::dp1200, RPS1/rps1Δ:: Clp10 | This work |
| HMY194 | As CAI4, but mnn47Δ::dp1200/mnn47Δ::dp1200, RPS1/rps1Δ:: Clp10 | This work |
| HMY195 | As CAI4, but mnn41Δ::dp1200/mnn41Δ::dp1200, mnn47Δ::dp1200/mnn47Δ::dp1200, RPS1/rps1Δ:: Clp10 | This work |
| HMY196 | As CAI4, but mnn41Δ::dp1200/mnn41Δ::dp1200, mnn47Δ::dp1200/mnn47Δ::dp1200, RPS1/rps1Δ:: Clp10-MNN41 | This work |
| HMY197 | As CAI4, but mnn41Δ::dp1200/mnn41Δ::dp1200, mnn47Δ::dp1200/mnn47Δ::dp1200, RPS1/rps1Δ:: Clp10-MNN47 | This work |
| HMY198 | As CAI4, but mnn41Δ::dp1200/mnn41Δ::dp1200, mnn42Δ::dp1200/mnn42Δ::dp1200, mnn47Δ::dp1200/mnn47Δ::dp1200, RPS1/rps1Δ:: Clp10 | This work |
| HMY199 | As CAI4, but mnn41Δ::dp1200/mnn41Δ::dp1200, mnn42Δ::dp1200/mnn42Δ::dp1200, mnn47Δ::dp1200/mnn47Δ::dp1200, RPS1/rps1Δ:: Clp10-MNN41 | This work |
| HMY200 | As CAI4, but mnn41Δ::dp1200/mnn41Δ::dp1200, mnn42Δ::dp1200/mnn42Δ::dp1200, mnn47Δ::dp1200/mnn47Δ::dp1200, RPS1/rps1Δ:: Clp10-MNN42 | This work |
| HMY201 | As CAI4, but mnn41Δ::dp1200/mnn41Δ::dp1200, mnn42Δ::dp1200/mnn42Δ::dp1200, mnn47Δ::dp1200/mnn47Δ::dp1200, RPS1/rps1Δ:: Clp10-MNN47 | This work |
| HMY202 | As CAI4, but mnn43Δ::dp1200/mnn43Δ::dp1200, mnn44Δ::dp1200/mnn44Δ::dp1200, mnn45Δ::dp1200/mnn45Δ::dp1200, mnn46Δ::dp1200/mnn46Δ::dp1200, RPS1/rps1Δ:: Clp10 | This work |
| NGY648 | As BWP17, but mnn4Δ/mnn4Δ, mnn41Δ/mnn41Δ, mnn42Δ/mnn42Δ, mnn43Δ/mnn43Δ, mnn44Δ/mnn44Δ, mnn45Δ/mnn45Δ, mnn46Δ/mnn46Δ, mnn47Δ/mnn47Δ | This work |
| HMY95 | As CDH7, but RPSI/rps1Δ::Clp10-MNN41 | This work |
| HMY92 | As CDH7, but RPSI/rps1Δ::Clp10-MNN42 | This work |
| HMY104 | As CDH7, but RPSI/rps1Δ::Clp10-MNN43 | This work |
| HMY167 | As CDH7, but RPSI/rps1Δ::Clp10-MNN44 | This work |
| HMY168 | As CDH7, but RPSI/rps1Δ::Clp10-MNN45 | This work |
| HMY164 | As CDH7, but RPSI/rps1Δ::Clp10-MNN46 | This work |
| HMY123 | As CDH7, but RPSI/rps1Δ::Clp10-MNN47 | This work |
| HMY96 | As NGY522, but RPS1/rps1Δ::CIp10-MNN41 | This work |
| HMY87 | As NGY522, but RPS1/rps1Δ::CIp10-MNN42 | This work |
| HMY103 | As NGY522, but RPS1/rps1Δ::CIp10-MNN43 | This work |
| HMY165 | As NGY522, but RPS1/rps1Δ::CIp10-MNN44 | This work |
| HMY159 | As NGY522, but RPS1/rps1Δ::CIp10-MNN45 | This work |
| HMY166 | As NGY522, but RPS1/rps1Δ::CIp10-MNN46 | This work |
| HMY124 | As NGY522, but RPS1/rps1Δ::CIp10-MNN47 | This work |
Construction of null mutants
The mini-ura-blaster technique was used for gene disruption as follows. Specific primer pairs for each gene, containing complementary sequences to the 5′- and 3′- regions of the target ORF (see Supplementary Material, Table S1) were used to amplify by PCR the disruption cassette from the pDDB57 plasmid (Wilson et al., 2000). The strain CAI4, a Ura− mutant derived from the clinical isolated SC5314 (Fonzi and Irwin, 1993), was transformed sequentially with the disruption cassettes and the URA3 marker recycled by growing transformants on SC medium supplemented with 1 mg/mL 5-fluoroorotic acid and uridine. The plasmid CIp10 was used to restore URA3 at the RPS1 locus, as previously described (Murad et al., 2000). For gene disruption using the CRISPR-Cas9 strategy, C. albicans strain BWP17 (Wilson et al., 1999) was used as parental strain. Plasmid for CaCas9 Solo system (pV1200 vector) was adopted in this work (Vyas et al., 2015). Constructions of knockout vectors, marker recycling and verification of CRISPR-mutagenized loci were performed according to the previous method (Vyas et al., 2015). The oligonucleotide sequences used in this study are listed in Table S2.
Generation of constructions to complement null mutant strains
The ORF of each MNN4-like gene-family, plus ~1,000 bp upstream and ~600 bp downstream were amplified by PCR using primers listed in Table S3. The PCR product was cloned into NotI sites of the CIp10 plasmid (Murad et al., 2000). The null mutants generated in this study, a mnn4Δ null strain (Hobson et al., 2004) and a double mnt3Δ, mnt5Δ null strain were transformed with the constructions generated (see Table 1). Before transformation, constructions were digested with StuI, and confirmation of plasmid insertion into the RPS1 locus was performed by PCR.
Expression analysis
Total RNA was isolated from yeast cells using TRIzol (Invitrogen), and further purified with the kit RNeasy (Qiagen), following the manufacture's instruction. The cDNA was synthesized using the SuperScript system (Invitrogen). Following synthesis, cDNA purification was conducted after RNA degradation using adsorption chromatography, as described (Trujillo-Esquivel et al., 2016). Absence of genomic DNA in the cDNA preparations was confirmed by amplification of the ACT1 gene which contains an intron of 658 bp (data not shown).
All primer pairs used in qPCR reactions are listed in Table S4. The reaction mixtures were prepared using the SYBR® Green PCR Master Mix (Thermo Fisher Scientific) and analyzed in a StepOnePlus Real-Time PCR System (Applied Biosystems). All reactions produced a single amplicon, with a uniform melting curve, as determined by the dissociation profile of the products. Relative quantification was determined by calculating 2−ΔΔCT (Livak and Schmittgen, 2001). Expression data were normalized using RPP2B, a housekeeping gene previously used as a control in expression assays (Nailis et al., 2006).
Alcian blue binding assays
The phosphomannan content was determined by measuring the binding of the cationic dye Alcian Blue to the cell surface, as previously described (Hobson et al., 2004). Briefly, cells in exponential growth phase were collected, washed twice with deionized water and adjusted to an OD600 of 0.2. Aliquots of 1 mL were pelleted, and cells suspended in 1 mL of 30 μg/mL Alcian Blue (in 0.02 M HCl) and incubated at room temperature for 15 min. The cell suspension was then centrifuged and the supernatant saved and used to quantify the content of Alcian Blue absorbance at 620 nm. The concentration of the free (non-bound) dye was then determined and used to calculate the amount of Alcian Blue bound to cells as described (Hobson et al., 2004).
Phenotypic characterization of null mutants
Cells from overnight cultures grown in YPD at 28°C and 200 rpm were used to inoculate fresh YPD broth and cell growth was monitored every 30 min by absorbance at 600 nm. Yeast-hypha dimorphism was evaluated by incubating 5 × 106 cells/mL in RPMI 1640 (Sigma) supplemented with 10% (v/v) fetal bovine serum (Sigma) for 4 h at 37°C and 200 rpm. Cell preparations were inspected by bright-field microscopy to evaluate the percentage of yeast cells, pseudohyphae and hyphae. Two hundred cells were counted per strain. Alternatively, hypha formation was stimulated by growing cells on solid Spider medium (Liu et al., 1994). The robustness of the cell wall was tested by assessing the sensitivity of yeast cells to a range of cell wall perturbing agents. Cells in the exponential growth phase were collected, washed twice with deionized water, adjusted to an OD600 of 0.05 and seeded into a 96-well plate containing YPD plus doubling dilutions of: SDS, Calcofluor White, Congo Red, and hygromycin B. Plates were incubated at 28°C for 24 h and then read at OD600. The highest concentrations tested for each agent were: 0.25% (v/v) SDS, 100 μg/mL Calcofluor White, 100 μg/mL Congo Red, and 500 μg/mL hygromycin B.
Phagocytosis analysis
The RAW 264.7 (ATCC® TIB-71™) murine cell line was cultured in DMEM media (Sigma), supplemented with 10% (v/v) fetal bovine, at 37°C and 5% (v/v) CO2. After reaching 90% confluence, cells were detached using trypsin (Sigma) in DMEM medium and sub-seeded into 6-well plates for further growth. Cells were again harvested by trypsin detachment and the concentration adjusted to 2 × 105 cells/mL in DMEM medium. Yeast cells were grown in YPD at 28°C with reciprocal shaking at 200 rpm until they reached exponential growth phase and were then washed twice with PBS, and labeled simultaneously with Acridine Orange (1 mg/mL, Sigma) as reported (Abrams et al., 1983). The fungal cells were washed twice with PBS and resuspended at a cell density of 1 × 107 cell/mL. Phagocytosis assays were carried out in a total volume of 800 μL of fresh DMEM, in 6-well plates. The macrophage:yeast cell ratio was set at 1:3, and interactions were incubated for 2.5 h at 37°C under a CO2 atmosphere. Macrophages were washed once with PBS and detached from plates using trypsin. Cells were washed twice with PBS by centrifuging at 200 × g for 10 min at 4°C, and resuspended in PBS containing 1.25 mg/mL Trypan Blue as an external fluorescence quencher, as described previously (Santos et al., 2015). Macrophages were kept on ice until they were analyzed by flow cytometry.
Flow cytometry was performed in a MoFlo XDP system (Beckman Coulter) collecting 50,000 events that were singlet events, and then gated for macrophage cells. Fluorescence was recovered from the compensated FL1 (green) and FL3 (red) channels using macrophage cells without any labeling. Phagocytosis of yeast cells was assessed by counts in the green (recently phagocytosed cells) and red (cells within acidified phagolysosomes) fluorescence channels.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 7 software. All the experiments were performed with three biological replicates in duplicate. Data represent cumulative results of all experiments performed and are shown as means ± S.D. The Mann-Whitney U-test was used to establish statistical significance, which was set at P < 0.05.
Results
Disruption of C. albicans MNN4 family genes
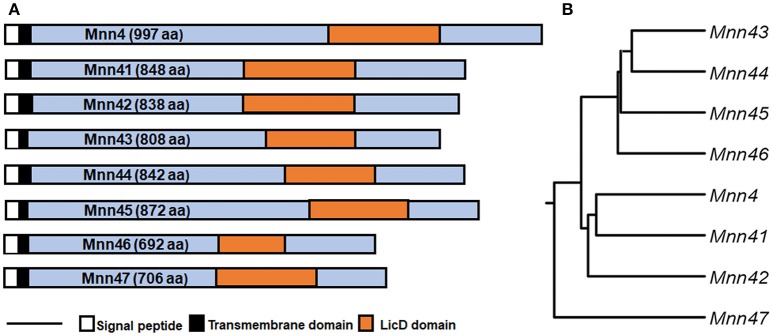
The C. albicans MNN4 gene has been previously characterized, and encodes a protein of 997 amino acids, with a putative signal peptide, a transmembrane domain, and an N-terminal region of lysine/glutamic acid repeats that is essential for enzyme activity (Hobson et al., 2004). Near the C-terminal end is a domain that is common to members of the LicD family of proteins, which are involved in phosphocholine metabolism (Zhang et al., 1999). This domain is present in proteins that belong to the nucleotidyltransferase superfamily, which includes enzymes that catalyze the transfer of nucleoside monophosphate to the hydroxyl group of an acceptor, using a nucleoside triphosphate as the donor (Kuchta et al., 2009). Bioinformatics analyses of putative encoded products of MNN4-like genes indicated that all of them have the LicD domain, a putative signal peptide, and a transmembrane domain close to the N-terminus (Figure 1A). These analyses did not identify lysine/glutamic acid repeats in the other MNN4 genes, but some revealed the presence of glutamine repeats, of 2–4 amino acids: For Mnn45 these repeats were closer to the N-terminus, whereas in Mnn41, Mnn42, and Mnn47 they were closer to the C- terminus. No glutamine repeats were identified in Mnn43, Mnn44, and Mnn46 (data not shown). All the seven family members had similarity scores of ~50% to Mnn4, varying between 43% for Mnn41 and Mnn45 to 58% for Mnn42 and Mnn47 (Table 2). The putative amino acid sequences of the MNN4-like genes were used to generate a phylogenetic tree (Figure 1B), using the “Neighbor-joining tree without distance correction” algorithm (Sievers et al., 2011). This analysis showed three main groups: one composed of Mnn43, Mnn44, Mnn45, and Mnn46; the second of Mnn4, Mnn41, and Mnn42; while Mnn47 was isolated as a single outlier (Figure 1B). This dendrogram was used to design subsequent sequential gene disruption strategies.
Figure 1.
Phylogenetic tree of members of the C. albicans MNN4-like gene family. (A) The eight members of the MNN4-like gene family, indicating relevant features of the primary amino acid sequences. The scale bar represents 100 amino acids (B), Dendrogram generated using the Neighbour-joining tree without distance correction algorithm defining three clades, (i) Mnn43, Mnn44, Mnn45 and Mnn46; (ii) Mnn4, Mnn41 and Mnn42; and (iii) Mnn47 as a sole outlier. The amino acid sequences were retrieved from http://www.candidagenome.org, archived under the following systematic names: MNN4 (C4_06540W_A), MNN41 (C2_03710W_A), MNN42 (C2_03690C_A), MNN43 (C1_02670C_A), MNN44 (C1_02680C_A), MNN45 (C6_02830W_A), MNN46 (C4_06990W_A), MNN47 (C1_09130W_A).
Table 2.
Comparison of the putative protein sequence of the C. albicans MNN4-like gene family members.
| Mnn4 | Mnn41 | Mnn42 | Mnn43 | Mnn44 | Mnn45 | Mnn46 | Mnn47 | |
|---|---|---|---|---|---|---|---|---|
| Mnn4 | 100/100* | 40/56 | 41/61 | 32/50 | 31/49 | 34/51 | 35/52 | 39/57 |
| Mnn41 | 40/56 | 100/100 | 34/51 | 29/44 | 30/48 | 29/43 | 28/46 | 29/46 |
| Mnn42 | 41/61 | 34/51 | 100/100 | 27/46 | 26/46 | 31/52 | 31/49 | 35/58 |
| Mnn43 | 32/50 | 29/44 | 27/46 | 100/100 | 36/56 | 33/52 | 31/48 | 26/46 |
| Mnn44 | 34/51 | 30/48 | 26/46 | 36/56 | 100/100 | 34/51 | 32/49 | 30/54 |
| Mnn45 | 32/49 | 29/43 | 31/52 | 33/52 | 34/51 | 100/100 | 32/48 | 35/55 |
| Mnn46 | 35/52 | 28/46 | 31/49 | 31/48 | 32/49 | 32/48 | 100/100 | 27/46 |
| Mnn47 | 39/57 | 29/46 | 35/58 | 26/46 | 30/54 | 35/55 | 27/46 | 100/100 |
Numbers represent percentage of identity and similarity, respectively.
The C. albicans strain CAI4, a Ura− derived strain from the clinical isolate SC5314 (Fonzi and Irwin, 1993), was used as genetic background for the generation of single null mutants, using the mini-Ura-blaster strategy (Wilson et al., 2000). These null mutants were used as background to generate double, triple, or quadruple null mutants. To avoid problems due to ectopic expression of URA3 (Brand et al., 2004), the resulting null mutants were transformed with the StuI-linearized CIp10 plasmid (Murad et al., 2000), introducing URA3 at the neutral RPS1 locus. The strain CAI4 transformed with the StuI-linearized CIp10 plasmid (NGY152) (Brand et al., 2004) was used as a wild-type (WT) control strain. The re-integrant control strains were constructed introducing the gene of interest at the RPS1 locus, as described in Materials and Methods. Furthermore, to generate a mutant strain lacking all eight MNN4-like genes, the CRISPR-Cas9 system (Vyas et al., 2015) was used in the BWP17 genetic background (Wilson et al., 1999). None of the strains generated in this study (see Table 1) displayed changes in the growth rate, ability to undergo dimorphism, cell and colony morphologies, and the sensitivity to the cell wall perturbing agents Congo Red, Calcofluor White, SDS, and hygromycin B (data not shown). Mutants derived independently in parallel lineages in which the order of disruption of individual genes differed, all generated the same final phenotype.
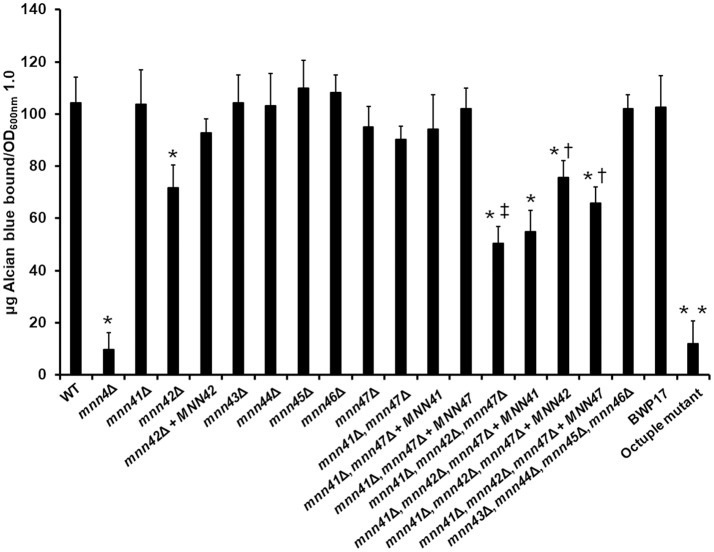
Next, we tested the ability of the single null mutants to bind Alcian Blue, a dye that specifically binds the negative charge provided by the phosphate group of phosphomannan (Hobson et al., 2004). The WT control cells efficiently bound Alcian Blue whilst the mnn4Δ was incapable of binding this cationic dye (Figure 2). Compared to the WT control strain, no significant changes were observed in the ability of the mnn41Δ, mnn43Δ, mnn44Δ, mnn45Δ, and mnn46Δ null mutants to bind the dye. However, the mnn42Δ null mutant displayed a significant reduction in Alcian Blue binding, which was restored in the re-integrant control strain (Figure 2). Although it was not significant at P > 0.05, the mnn47Δ null mutant showed a reduction in the ability to bind the dye (P = 0.27). Since Figure 1 indicates Mnn41, Mnn42, and Mnn47 are closest paralogs to Mnn4, and because Mnn42 is required for full cell wall phosphomannosylation (Figure 2), we then generated a double null mutant lacking both MNN41 and MNN47. This double mutant showed binding levels comparable to those found in the WT control strain, indicating MNN42 was enough to sustain the normal cell wall phosphomannosylation (Figure 2). A triple mnn41Δ, mnn42Δ, and mnn47Δ null mutant was significantly attenuated in the ability to bind Alcian Blue compared to the WT control cells or the single mnn42Δ null mutant (Figure 2), indicating these family members may operate in tandem in cell wall phosphomannosylation. Complementation of this strain with MNN41 did not increase Alcian Blue binding, but complementation with either MNN42 or MNN47, resulted in a significant elevation in Alcian Blue binding in the triple null mutant (Figure 2), suggesting that MNN42 and MNN47 can play a significant role in the C. albicans cell wall phosphomannosylation. Mnn43, Mnn44, Mnn45, and Mnn46 belong to a different clade of the Mnn4 family (Figure 1). Therefore, we generated a quadruple null mutant lacking all four of these genes. However, this quadruple mutant did not show significant changes in the ability to bind Alcian Blue. The CRISPR-Cas9 single mutants reproduced the same Alcian Blue patterns generated by mini-Ura-Blaster disruption (data not shown). The CRISPR Cas9 generated octuple mutant, lacking all the MNN4-like genes and MNN4 was unable to bind Alcian Blue, showing a phenotype similar to that observed in the mnn4Δ null mutant (Figure 2). These data indicate that some, but not all the members of the MNN4-like gene family participate in the C. albicans cell wall phosphomannosylation, and confirmed the dominant role of MNN4 during phosphomannan synthesis in cells grown under standard laboratory conditions.
Figure 2.
Members of the C. albicans MNN4-like gene family participate in cell wall phosphomannosylation. Cells were grown in YPD medium, and ability to bind Alcian Blue was measured. The strains used are: NGY152 (WT), CDH15 (mnn4Δ), HMY189 (mnn41Δ), HMY81 (mnn42Δ), HMY84 (mnn42Δ + MNN42), HMY190 (mnn43Δ), HMY191 (mnn44Δ), HMY192 (mnn45Δ), HMY193 (mnn46Δ), HMY194 (mnn47Δ), HMY195 (mnn41Δ, mnn47Δ), HMY196 (mnn41Δ, mnn47Δ + MNN41), HMY197 (mnn41Δ, mnn47Δ + MNN47), HMY198 (mnn41Δ, mnn42Δ, mnn47Δ), HMY199 (mnn41Δ, mnn42Δ, mnn47Δ + MNN41), HMY200 (mnn41Δ, mnn42Δ, mnn47Δ + MNN42), HMY201 (mnn41Δ, mnn42Δ, mnn47Δ + MNN47), HMY202 (mnn43Δ, mnn44Δ, mnn45Δ, mnn46Δ); BWP17, and NGY648 (octuple mutant).The data represent the means ± SD of three independent assays performed by triplicate. *P < 0.05 when compared against the WT strain. †P < 0.05 when compared against the mnn41Δ, mnn42Δ, mnn47Δ null mutant. ‡P < 0.05 for the comparison of the triple mnn41Δ, mnn42Δ, mnn47Δ, and the mnn42Δ null mutant. **P < 0.05 when the octuple mutant was compared against the parental strain BWP17.
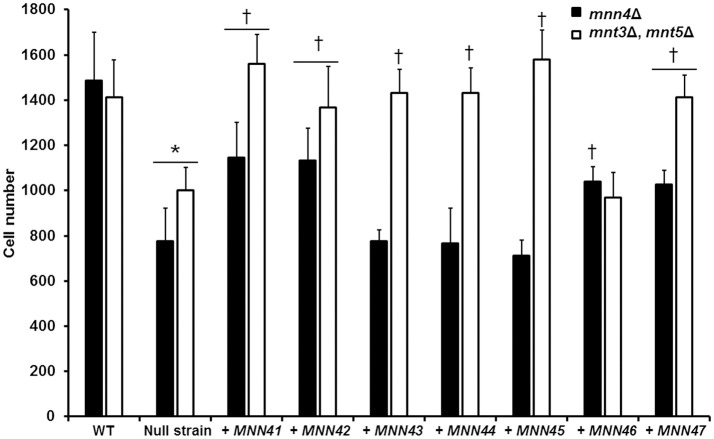
Incremental increases in the gene dose of either MNN41, MNN42, MNN46, or MNN47 partially complement the C. albicans MNN4Δ null mutant
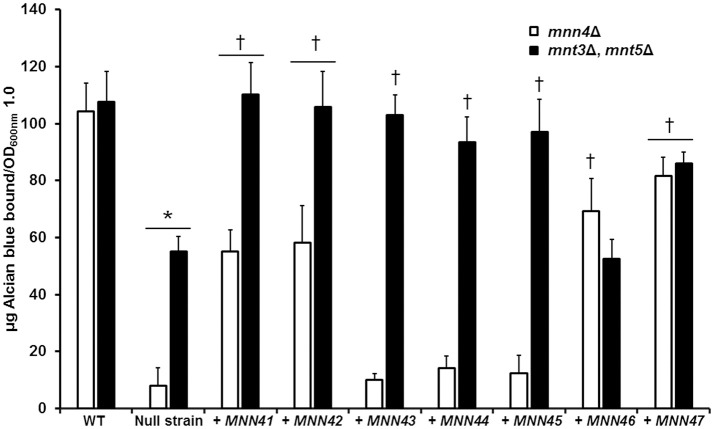
We next performed experiments in which transformed additional alleles of each member of the MNN4-like gene family into mutant strains lacking a functional MNN4, to assess whether this impacted on the ability to bind Alcian Blue. A Ura− strain lacking MNN4 (strain CDH7, see Table 1) was transformed with the StuI-linearized CIp10 plasmid (Murad et al., 2000) harboring alleles of the MNN4-like gene family, generating a series of plasmids with an extra copy of each of these genes (see Table 1). All of these genes were expressed under the control of their own regulatory upstream sequences. We first analyzed whether the resulting increase in gene dosage impacted the expression levels of other MNN4 genes. It was shown (Table 3) that in the mnn4Δ null mutant background the expression of other family members was not significantly affected with two exceptions - MNN42 expression almost doubled and MNN47 expression was increased by about 70%. All strains showed a significantly increased expression of the supplemented gene, when extra copies were transformed into the mnn4Δ null mutant background (Table 3). Therefore, incremental increases in the gene dose had a positive impact on the expression level of each member of the MNN4-like gene family. Next, we measured the ability of these strains to bind Alcian Blue and found that none of the complemented transformants were capable of restoring wild-type levels of Alcian Blue binding (Figure 3). However, the incremental increase in the gene dosage of MNN41, MNN42, MNN46, or MNN47 resulted in strains that had significantly higher levels of dye bound, compared to the mnn4Δ null mutant (Figure 3). Therefore, increased expression of some, but all MNN4-like genes, was capable of partially complementing the mnn4Δ null mutant.
Table 3.
Analysis of the expression of the members of the C. albicans MNN4-like gene family.
| Gene | WT* | mnn4Δ† | mnn4Δ + EV ‡ | mnt3Δ, mnt5Δ¶ | mnt3Δ, mnt5Δ + EV ‡ |
|---|---|---|---|---|---|
| MNN41 | 1.0 ± 0.01 | 1.1 ± 0.2 | 1.7 ± 0.1†† | 1.0 ± 0.09 | 1.8 ± 0.3†† |
| MNN42 | 1.0 ± 0.01 | 1.9 ± 0.1** | 2.5 ± 0.2†† | 1.1 ± 0.1 | 1.9 ± 0.1†† |
| MNN43 | 1.0 ± 0.03 | 1.1 ± 0.2 | 1.8 ± 0.1†† | 1.0 ± 0.05 | 1.7 ± 0.06†† |
| MNN44 | 1.0 ± 0.02 | 1.0 ± 0.1 | 2.0 ± 0.4†† | 1.1 ± 0.1 | 1.8 ± 0.1†† |
| MNN45 | 1.0 ± 0.01 | 1.1 ± 0.3 | 1.9 ± 0.1†† | 1.1 ± 0.1 | 1.8 ± 0.09†† |
| MNN46 | 1.0 ± 0.03 | 1.0 ± 0.2 | 1.9 ± 0.3†† | 1.0 ± 0.08 | 1.7 ± 0.1†† |
| MNN47 | 1.0 ± 0.02 | 1.7 ± 0.2** | 2.6 ± 0.3†† | 1.1 ± 0.1 | 1.9 ± 0.05†† |
Strain NGY152.
Strain CDH15.
EV, Expression vector, see Table 1 for strain details.
Strain NGY1227.
P < 0.05, when compared to the WT strain.
P < 0.05, when compared to the null mutant strain.
Figure 3.
Incremental increases in the allele number of members of the C. albicans MNN4-like gene family restores the ability to bind Alcian Blue in mutants with defects in the phosphomannosylation. Null mutant cells lacking either mnn4Δ (open bars) or mnt3Δ, mnt5Δ (closed bars) were transformed with expression vectors containing each of the members of the MNN4-like gene family, then were grown in YPD medium, and ability to bind Alcian Blue was measured. The strains used are: NGY152 (WT), CDH15 (mnn4Δ), NGY1227 (mnt3Δ, mnt5Δ), see Table 1 for details of other strains. The data represent the means ± SD of three independent assays performed by triplicate. *P < 0.05, compared with WT control cells. †P < 0.05 when compared to the null mutant strain. The open bars correspond to strains in which the mnn4Δ null mutant was complemented with MNN4 gene alleles. The closed bars refer to experiments in which the double mnt3Δ, mnt5Δ null mutant was complemented.
Increment in the gene dose of MNN46 does not complement the C. albicans mnt3Δ, mnt5Δ null mutant
Using the same strategy, complementation experiments were performed using the double mnt3Δ, mnt5Δ null mutant background, which lacks the phosphomannosyltransferases required for 50% of the normal level of total cell wall phosphomannan (Mora-Montes et al., 2010). The Ura− strain carrying this dual disruption was used as the parent for complementing transformations with all individual MNN4-like genes (Table 1). Expression analyses indicated that loss of both MNT3 and MNT5 did not affect the expression of other MNN4-like gene family members (Table 3). Upon insertion of an additional MNN4 allele, the expression of the relevant family member was significantly increased (Table 3). When extra copies of the genes encoding MNN41, MNN42, MNN43, MNN44, MNN45, or MNN47 were introduced in the double mnt3Δ, mnt5Δ null background, the resulting increased expression of each gene increased the ability of the double null mutant to bind Alcian Blue (Figure 3). However, increased MNN46 expression in this double mutant did not affect Alcian Blue binding (Figure 3).
Alterations in the gene dosage of MNN4-like genes affects the C. albicans-macrophage interaction
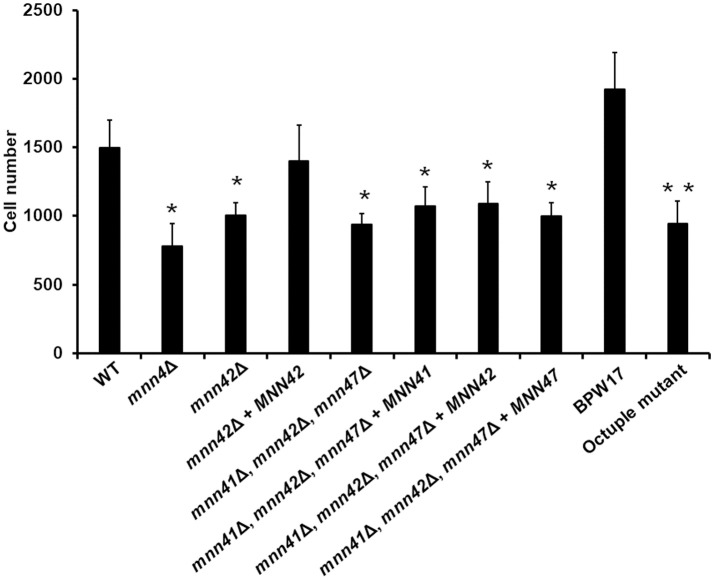
Because disruption of the cell wall phosphomannosylation is related to defective phagocytosis of C. albicans by macrophages (McKenzie et al., 2010), we then assessed whether gene disruption or gene supplementation of the MNN4-like gene family members impacted C. albicans-macrophages interactions. As reported previously (McKenzie et al., 2010), the mnn4Δ null mutant strain exhibited a 50% reduction in phagocytosis compared to WT control cells (Figures 4, 5). The mnn42Δ null mutant also showed a significant reduction in uptake by macrophages, that was restored to WT levels in the re-integrant control strain (Figure 4). The triple mnn41Δ, mnn42Δ, mnn47Δ null mutant also displayed a reduction in the phagocytosis when interacting with human macrophages, but reintroduction of a single disrupted gene failed to complement this phenotype (Figure 4). Although the parental strain BPW17 displayed increased phagocytosis compared to the NGY52 (WT) control strain (P = 0.0932, when compared WT and BPW17), the octuple null mutant showed a 50% reduction in the uptake by macrophages (Figure 4). The other null mutant strains generated in this work were phagocytosed to a similar extent to WT control cells (data dot shown).
Figure 4.
Phagocytosis of C. albicans null mutants lacking members of the MNN4-like gene family by RAW 264.7 macrophages. C. albicans yeast cells were labeled with Acridine Orange and incubated with RAW 264.7 macrophages at a MOI 3:1 for 2.5 h at 37°C under CO2. Macrophages were gated by FACS and 50,000 cells were counted/ sample. Results represent macrophages interacting with at least one fluorescent fungal cell. Strains used are: NGY152 (WT), CDH15 (mnn4Δ), HMY81 (mnn42Δ), HMY84 (mnn42Δ + MNN42), HMY198 (mnn41Δ, mnn42Δ, mnn47Δ), HMY199 (mnn41Δ, mnn42Δ, mnn47Δ + MNN41), HMY200 (mnn41Δ, mnn42Δ, mnn47Δ + MNN42), and HMY201 (mnn41Δ, mnn42Δ, mnn47Δ + MNN47). The data represent means ± SD for three independent assays performed by duplicate. *P < 0.05 when compared against the WT strain. **P < 0.05 when the octuple mutant was compared against the parental strain BWP17.
Figure 5.
Transformation of the mnn4 null mutant with additional alleles of the C. albicans MNN4-like gene family restores the C. albicans-RAW 264.7 macrophage interaction. Null mutant cells lacking either mnn4Δ (closed bars) or mnt3Δ, mnt5Δ (open bars) were transformed with expression vectors containing each of the members of the MNN4-like gene family. Cells were labeled with Acridine Orange and incubated with RAW 264.7 macrophages at a MOI 3:1 for 2.5 h at 37°C under a CO2 atmosphere. Then, macrophages were gated by FACS system and 50,000 cells were counted/sample. Results represent macrophages interacting with at least one fluorescent fungal cell. Strains used are: NGY152 (WT), CDH15 (mnn4Δ), NGY1227 (mnt3Δ, mnt5Δ), see Table 1 for details of the other strains. The data represent means ± SD of three independent biological replicates performed by duplicate. *P < 0.05, compared with WT control cells. †P < 0.05 is a comparison with the null mutant strain.
When the interaction of macrophages and cells harboring an extra copy of the members of the MNN4-like gene family was analyzed, we observed that the cell uptake followed a similar trend to that described in the ability of binding Alcian Blue. Extra copies of either MNN41, MNN42, or MNN46 in the mnn4Δ null background significantly increased yeast cell phagocytosis (Figure 5). Extra copies of all the family members, with the exception of MNN46, positively influenced the phagocytosis of cells in a genetic background lacking both MNT3 and MNT5 (Figure 5). Collectively, these data indicate the members of the MNN4-like gene family influence the C. albicans-human macrophage interaction and that phosphomannan content correlated positively with the degree of phagocytosis of the yeast cells by macrophages.
Discussion
The study of the fungal cell wall of medically relevant organisms is of special interest, because most cell wall components contribute to the induced immune response, and are required for cell fitness and virulence (Díaz-Jiménez et al., 2012; Martinez-Alvarez et al., 2014; Netea et al., 2015). Such studies have the potential to unveil new molecular targets to expand the repertoire of drugs to treat fungal infections. Cell wall phosphomannosylation is a glycoprotein modification found only in a reduced group of yeast-like fungi, including S. cerevisiae, Kloeckera brevis, Yarrowia lypolytica and several species of the Candida genus, including C. albicans (Jigami and Odani, 1999; Butler et al., 2009; Gil et al., 2015). Although, in C. albicans, this cell wall modification is dispensable for cell viability, it participates in the interaction with effector molecules and cells of the immune response (Harris et al., 2009; McKenzie et al., 2010), and is a trait that varies between clinical isolates, with some showing reduced and others increased phosphomannan content of their cell walls (MacCallum et al., 2009). This suggests cell wall phosphomannosylation is part of the cellular phenotype that C. albicans can modify as it adapts to different environments.
The molecular machinery resulting in the synthesis of phosphomannan is more complex in C. albicans than that described in S. cerevisiae, where all the components of this biosynthetic process have been analyzed. This complexity in C. albicans is underlined by the presence of an extended MNN4-like gene family. None of the other C. albicans MNN4-like genes contains the lysine/glutamic acid repeats found in Mnn4, which is presumed to be required for phosphomannan production (Hobson et al., 2004). This may offer an explanation for the dominant role of Mnn4, since deletion of this gene was sufficient to eliminate Alcian Blue binding of cells grown under laboratory conditions. A conserved feature of Mnn4 and Mnn4-family proteins is the presence of the LicD domain. The C. albicans genome database identified only 8 encoded proteins containing this domain - Mnn4 and the seven MNN4-like family members. A similar analysis within the S. cerevisiae genome database identified only Mnn4 and Mnn14 as proteins containing this domain (data not shown). Although the functional role of this LicD domain in the phosphomannosylation mechanism remains unknown, we conclude this is a signature domain for the identification of proteins involved in the cell wall phosphomannosylation pathway in fungi. In Ogataea minuta (Akeboshi et al., 2009), Yarrowia lipolytica (Park et al., 2011), Pichia pastoris (Miura et al., 2004), Candida parapsilosis, and Candida tropicalis (our unpublished observations), the Mnn4-like proteins involved in the modification of glycoproteins with phosphomannan all contain the LiCD domain, providing additional support for this conclusion.
The null mutants generated here displayed no defects in morphology, ability to undergo yeast-hypha dimorphism and sensitivity to a range of cell wall perturbing agents. This is consistent with the phenotype reported for the mnn4Δ null mutant (Hobson et al., 2004). The only measured alteration in the phenotypes was the ability of cells to bind Alcian Blue and to interact with macrophages (Hobson et al., 2004; McKenzie et al., 2010). However, we would predict that any phenotype that depends on the net charge of the cell wall would likely to be affected, including the interaction with cationic peptides. Even though most of the single null mutants generated here did not show a marked reduction in the cell wall phosphomannan content, the subtle changes in phosphomannan content still affected the interaction with macrophages, indicating that, along with mannan and β1, 3-glucan (Heinsbroek et al., 2008), phosphomannan is a key cell wall component influencing immune recognition. Interestingly, although the cell wall phosphomannan content of the mnn41, mnn42, mnn47 triple null mutant was partially restored by complementation with either MNN42 or MNN47, this did not have a significant impact on the phagocytosis of the complemented strains. These data suggest that a minimal concentration of phosphomannan has to be present in the wall for optimal rates of phagocytosis by human macrophages. Since the full reduction of cell wall phosphomannan in the mnn4Δ null mutant did not affect virulence in the murine model of candidiasis (Hobson et al., 2004), we did not perform an exhaustive analysis of the virulence properties of this mutant collection.
The generation of an octuple mutant in C. albicans using the recently developed CRISPR-Cas9 system (Vyas et al., 2015) is one of the most extensive disruption protocols attempted in this fungus. To our knowledge, the disruption of a full gene family has been only reported for the KRE2/MNT1 and MNN2 gene families, which are composed of five and six members, respectively (Mora-Montes et al., 2010; Hall et al., 2013). The octuple mutant reported demonstrates the advantages that the CRISPR-Cas9 strategy can provide in the assessment of extended gene families in this organism.
We demonstrate that Mnn42, and to a lesser extent Mnn47, were the MNN4 family members (other and Mnn4) that most affected cell wall phosphomannan synthesis. These genes were upregulated in a genetic background lacking MNN4. These data suggest that Mnn42 and Mnn47 contribute to canonical glycoprotein modification with mannosylphosphate, but that the presence of Mnn4 is obligately required to carry out this function. When the gene dose of MNN4-like gene family members was increased by transformation, we observed that overexpression of MNN41, MNN42, MNN46 and MNN47 partially restored the cell wall phosphomannan, indicating that these genes can compensate for the absence of Mnn4, only when their expression levels were increased. These findings support studies in C. albicans and S. cerevisiae, where strains that were heterozygous for the MNN4 locus were unable to bind normal levels of Alcian Blue (Jigami and Odani, 1999; Hobson et al., 2004). We found that the transcript abundance was similar for MNN4 and the MNN4-like gene family members in the WT control strain when cells were grown in YPD at 28°C (data not shown). Therefore, the dominant contribution of MNN4 to phosphomannosylation over the members of the gene family may rely on posttranscriptional or posttranslational modifications that favor the accumulation of Mnn4 in the system. Analysis of the public available databases analyzed via transcriptomic microarrays revealed that MNN4, MNN43, and MNN45 only undergo subtly altered levels of transcription when cells were grown under a variety of conditions, or exposed to different stressors (Enjalbert et al., 2003, 2006; Lorenz et al., 2004; Fradin et al., 2005; Karababa et al., 2006). However, expression of MNN41 and MNN42 was shown to be upregulated when the fungal cells interact with neutrophils (Fradin et al., 2005). The transcriptional regulation of the latter is also positively influenced when cells are grown in presence of 200 mM CaCl2 for 20, 40, or 60 min (Karababa et al., 2006). Expression of MNN44 and MNN46 was reported to be downregulated and upregulated, respectively, when cells were incubated under oxidative stress conditions (Enjalbert et al., 2003). Furthermore, MNN46 expression was upregulated as a response to nitrogen or carbon starvation (Lorenz et al., 2004). Collectively, these results indicate the members of this gene family are transcriptionally regulated under a variety of growth conditions and that this regulation may influence the degree of glycoprotein phosphomannosylation. Moreover, it is not clear how members of this gene family are regulated in different micro niches during the natural history of an infection.
Overexpression of MNN4-like gene family members in a genetic background lacking Mnt3 and Mnt5 (Mora-Montes et al., 2010) also demonstrated that the gene dosage of any of the family members, with the exception of MNN46, restored the phosphomannan content to levels comparable to those found in the WT control cells. Again, this suggests that activation of phosphomannosyltransferase could be achieved by proteins that are solely activated by Mnn4, or proteins activated by the dual action of Mnn4 and other MNN4-like gene family members. This reinforces and extends previous reports studying the canonical Mnn4 protein (Hobson et al., 2004). This conclusion is inferred from the current report, where overexpression of most of the family members positively affected the phosphomannosylation thereby partially compensating for the lack of Mnn4. It is formally possible that all MNN4-like gene family members participate directly as phosphomannosyltransferases. Additional experiments using in vitro assays for this biochemical reaction would be required to address this possibility unequivocally.
The data reported here clearly demonstrated that MNN4-like genes have a significant role in C. albicans phagocytosis by macrophages; which contrast with previously reported results from our group, indicating that a mnn4Δ null mutant was readily phagocytosed as the WT control cells (Hobson et al., 2004). McKenzie et al. (2010) reported a comprehensive study of the role of protein mannosylation during phagocytosis and found that the experimental setting published in 2004 was overshadowing the differences between the WT and mnn4Δ strains. Hobson et al. (2004) used a yeast:macrophage ratio 20:1, for 1 h at 37°C, whereas McKenzie et al. (2010), and this report, used a yeast:macrophage ratio 3:1 and longer incubation times (3 and 2.5 h, respectively). These technical differences are likely to account for the apparent contradiction between our observations and those previously reported (Hobson et al., 2004).
In conclusion, we provide one of the most comprehensive analyses of an extended gene family to be attempted in any human pathogen. We provide evidence that the eight members of the MNN4-like gene family members all are capable of participating in the synthesis of cell wall phosphomannan in C. albicans, although the majority of the cell wall phosphomannosylation, under normal growth conditions, is achieved via Mnn4 itself. In addition, the observation that deletion of all individual members of this family influenced the ability of macrophages to phagocytose C. albicans cells suggests that this protein family collaborates to generate the charge in the cell wall, which in turn affects a number of immune recognition functions. Our data also underline the fact that the charge on the cell wall is critically important for the process of phagocytosis of this group of fungal pathogens.
Author contributions
NG and HM conceived the study; RG, KJ, MH, ET, DC, AT, and BF performed the experiments; RG, KJ, MH, DD, ET, DC, AT, BF, NG, and HM analyzed the data; HM drafted the paper; RG, KJ, MH, DD, ET, DC, AT, BF, NG, and HM approved the final version of the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Luz A. López-Ramírez (Universidad de Guanajuato) for technical assistance. This work was supported by Consejo Nacional de Ciencia y Tecnología (ref. CB2011/166860; PDCPN2014-247109, and FC 2015-02-834), Universidad de Guanajuato (ref. 000025/11; 0087/13; ref. 1025/2016; Convocatoria Institucional para Fortalecer la Excelencia Académica 2015; CIFOREA 89/2016), Programa de Mejoramiento de Profesorado (ref. UGTO-PTC-261), and Red Temática Glicociencia en Salud (CONACYT-México). NG acknowledges the Wellcome Trust (086827, 075470, 101873, and 200208) and MRC Centre for Medical Mycology for funding (N006364/1). KJ was supported by a research visitor grant to Aberdeen from China Scholarship Council (CSC No. 201406055024).
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02156/full#supplementary-material
References
- Abrams W. R., Diamond L. W., Kane A. B. (1983). A flow cytometric assay of neutrophil degranulation. J. Histochem. Cytochem. 31, 737–744. 10.1177/31.6.6404983 [DOI] [PubMed] [Google Scholar]
- Akeboshi H., Kasahara Y., Tsuji D., Itoh K., Sakuraba H., Chiba Y., et al. (2009). Production of human β-hexosaminidase A with highly phosphorylated N-glycans by the overexpression of the Ogataea minuta MNN4 gene. Glycobiology 19, 1002–1009. 10.1093/glycob/cwp080 [DOI] [PubMed] [Google Scholar]
- Bain J. M., Louw J., Lewis L. E., Okai B., Walls C. A., Ballou E. R., et al. (2014). Candida albicans hypha formation and mannan masking of β-glucan inhibit macrophage phagosome maturation. MBio 5:e01874. 10.1128/mBio.01874-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates S., Hall R. A., Cheetham J., Netea M. G., MacCallum D. M., Brown A. J., et al. (2013). Role of the Candida albicans MNN1 gene family in cell wall structure and virulence. BMC Res. Notes 6:294. 10.1186/1756-0500-6-294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates S., Hughes H. B., Munro C. A., Thomas W. P., MacCallum D. M., Bertram G., et al. (2006). Outer chain N-glycans are required for cell wall integrity and virulence of Candida albicans. J. Biol. Chem. 281, 90–98. 10.1074/jbc.M510360200 [DOI] [PubMed] [Google Scholar]
- Bates S., MacCallum D. M., Bertram G., Munro C. A., Hughes H. B., Buurman E. T., et al. (2005). Candida albicans Pmr1p, a secretory pathway P-type Ca2+/Mn2+-ATPase, is required for glycosylation and virulence. J. Biol. Chem. 280, 23408–23415. 10.1074/jbc.M502162200 [DOI] [PubMed] [Google Scholar]
- Brand A., MacCallum D. M., Brown A. J., Gow N. A., Odds F. C. (2004). Ectopic expression of URA3 can influence the virulence phenotypes and proteome of Candida albicans but can be overcome by targeted reintegration of URA3 at the RPS10 locus. Eukaryot. Cell 3, 900–909. 10.1128/EC.3.4.900-909.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown G. D., Denning D. W., Gow N. A., Levitz S. M., Netea M. G., White T. C. (2012). Hidden killers: human fungal infections. Sci. Transl. Med. 4:165rv113. 10.1126/scitranslmed.3004404 [DOI] [PubMed] [Google Scholar]
- Butler G., Rasmussen M. D., Lin M. F., Santos M. A., Sakthikumar S., Munro C. A., et al. (2009). Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459, 657–662. 10.1038/nature08064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courjol F., Jouault T., Mille C., Hall R., Maes E., Sendid B., et al. (2015). Beta-1,2-mannosyltransferases 1 and 3 participate in yeast and hyphae O- and N-linked mannosylation and alter Candida albicans fitness during infection. Open Forum Infect. Dis. 2:ofv116. 10.1093/ofid/ofv116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Jimenez D. F., Mora-Montes H. M., Hernandez-Cervantes A., Luna-Arias J. P., Gow N. A., Flores-Carreon A. (2012). Biochemical characterization of recombinant Candida albicans mannosyltransferases Mnt1, Mnt2 and Mnt5 reveals new functions in O- and N-mannan biosynthesis. Biochem. Biophys. Res. Commun. 419, 77–82. 10.1016/j.bbrc.2012.01.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Jiménez D. F., Pérez-García L. A., Martínez-Álvarez J. A., Mora-Montes H. M. (2012). Role of the fungal cell wall in pathogenesis and antifungal resistance. Curr. Fungal Infect. Rep. 6, 275–282. 10.1007/s12281-012-0109-7 [DOI] [Google Scholar]
- Enjalbert B., Nantel A., Whiteway M. (2003). Stress-induced gene expression in Candida albicans: absence of a general stress response. Mol. Biol. Cell 14, 1460–1467. 10.1091/mbc.E02-08-0546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjalbert B., Smith D. A., Cornell M. J., Alam I., Nicholls S., Brown A. J. P., et al. (2006). Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol. Biol. Cell 17, 1018–1032. 10.1091/mbc.E05-06-0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwig L. P., Gow N. A. R. (2016). Interactions of fungal pathogens with phagocytes. Nat. Rev. Microbiol. 14, 163–176. 10.1038/nrmicro.2015.21 [DOI] [PubMed] [Google Scholar]
- Fonzi W. A., Irwin M. Y. (1993). Isogenic strain construction and gene mapping in Candida albicans. Genetics 134, 717–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin C., De Groot P., MacCallum D., Schaller M., Klis F., Odds F. C., et al. (2005). Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol. Microbiol. 56, 397–415. 10.1111/j.1365-2958.2005.04557.x [DOI] [PubMed] [Google Scholar]
- Gil J. Y., Park J. N., Lee K. J., Kang J. Y., Kim Y. H., Kim S., et al. (2015). Increased mannosylphosphorylation of N-glycans by heterologous expression of YlMPO1 in glyco-engineered Saccharomyces cerevisiae for mannose-6-phosphate modification. J. Biotechnol. 206, 66–74. 10.1016/j.jbiotec.2015.04.007 [DOI] [PubMed] [Google Scholar]
- Gow N. A. R., Hube B. (2012). Importance of the Candida albicans cell wall during commensalism and infection. Curr. Opin. Microbiol. 15, 406–412. 10.1016/j.mib.2012.04.005 [DOI] [PubMed] [Google Scholar]
- Hall R. A., Bates S., Lenardon M. D., Maccallum D. M., Wagener J., Lowman D. W., et al. (2013). The Mnn2 mannosyltransferase family modulates mannoprotein fibril length, immune recognition and virulence of Candida albicans. PLoS Pathog. 9:e1003276. 10.1371/journal.ppat.1003276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall R. A., Gow N. A. (2013). Mannosylation in Candida albicans: role in cell wall function and immune recognition. Mol. Microbiol. 90, 1147–1161. 10.1111/mmi.12426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris M., Mora-Montes H. M., Gow N. A., Coote P. J. (2009). Loss of mannosylphosphate from Candida albicans cell wall proteins results in enhanced resistance to the inhibitory effect of a cationic antimicrobial peptide via reduced peptide binding to the cell surface. Microbiology 155(Pt 4), 1058–1070. 10.1099/mic.0.026120-0 [DOI] [PubMed] [Google Scholar]
- Heinsbroek S. E., Taylor P. R., Martinez F. O., Martinez-Pomares L., Brown G. D., Gordon S. (2008). Stage-specific sampling by pattern recognition receptors during Candida albicans phagocytosis. PLoS Pathog. 4:e1000218. 10.1371/journal.ppat.1000218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobson R. P., Munro C. A., Bates S., MacCallum D. M., Cutler J. E., Heinsbroek S. E., et al. (2004). Loss of cell wall mannosylphosphate in Candida albicans does not influence macrophage recognition. J. Biol. Chem. 279, 39628–39635. 10.1074/jbc.M405003200 [DOI] [PubMed] [Google Scholar]
- Jigami Y., Odani T. (1999). Mannosylphosphate transfer to yeast mannan. Biochim. Biophys. Acta 1426, 335–345. 10.1016/S0304-4165(98)00134-2 [DOI] [PubMed] [Google Scholar]
- Karababa M., Valentino E., Pardini G., Coste A. T., Bille J., Sanglard D. (2006). CRZ1, a target of the calcineurin pathway in Candida albicans. Mol. Microbiol. 59, 1429–1451. 10.1111/j.1365-2958.2005.05037.x [DOI] [PubMed] [Google Scholar]
- Kim Y. H., Kang J.-Y., Gil J. Y., Kim S.-Y., Shin K. K., Kang H. A., et al. (2017). Abolishment of N-glycan mannosylphosphorylation in glyco-engineered Saccharomyces cerevisiae by double disruption of MNN4 and MNN14 genes. Appl. Microbiol. Biotechnol. 101, 2979–2989. 10.1007/s00253-017-8101-3 [DOI] [PubMed] [Google Scholar]
- Klis F. M., de Groot P., Hellingwerf K. (2001). Molecular organization of the cell wall of Candida albicans. Med. Mycol. 39(Suppl. 1), 1–8. 10.1080/744118876 [DOI] [PubMed] [Google Scholar]
- Kuchta K., Knizewski L., Wyrwicz L. S., Rychlewski L., Ginalski K. (2009). Comprehensive classification of nucleotidyltransferase fold proteins: identification of novel families and their representatives in human. Nucleic Acids Res. 37, 7701–7714. 10.1093/nar/gkp854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis L. E., Bain J. M., Lowes C., Gillespie C., Rudkin F. M., Gow N. A., et al. (2012). Stage specific assessment of Candida albicans phagocytosis by macrophages identifies cell wall composition and morphogenesis as key determinants. PLoS Pathog. 8:e1002578. 10.1371/journal.ppat.1002578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Kohler J., Fink G. (1994). Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266, 1723–1726. [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lorenz M. C., Bender J. A., Fink G. R. (2004). Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot. Cell 3, 1076–1087. 10.1128/EC.3.5.1076-1087.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum D. M., Castillo L., Nather K., Munro C. A., Brown A. J., Gow N. A., et al. (2009). Property differences among the four major Candida albicans strain clades. Eukaryot. Cell 8, 373–387. 10.1128/EC.00387-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Alvarez J. A., Perez-Garcia L. A., Flores-Carreon A., Mora-Montes H. M. (2014). The immune response against Candida spp. and Sporothrix schenckii. Rev. Iberoam. Micol. 31, 62–66. 10.1016/j.riam.2013.09.015 [DOI] [PubMed] [Google Scholar]
- Martinez-Duncker I., Diaz-Jimenez D. F., Mora-Montes H. M. (2014). Comparative analysis of protein glycosylation pathways in humans and the fungal pathogen Candida albicans. Int. J. Microbiol. 2014:267497. 10.1155/2014/267497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie C. G., Koser U., Lewis L. E., Bain J. M., Mora-Montes H. M., Barker R. N., et al. (2010). Contribution of Candida albicans cell wall components to recognition by and escape from murine macrophages. Infect. Immun. 78, 1650–1658. 10.1128/IAI.00001-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura M., Hirose M., Miwa T., Kuwae S., Ohi H. (2004). Cloning and characterization in Pichia pastoris of PNO1 gene required for phosphomannosylation of N-linked oligosaccharides. Gene 324, 129–137. 10.1016/j.gene.2003.09.023 [DOI] [PubMed] [Google Scholar]
- Mora-Montes H. M., Bates S., Netea M. G., Castillo L., Brand A., Buurman E. T., et al. (2010). A multifunctional mannosyltransferase family in Candida albicans determines cell wall mannan structure and host-fungus interactions. J. Biol. Chem. 285, 12087–12095. 10.1074/jbc.M109.081513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-Montes H. M., Bates S., Netea M. G., Diaz-Jimenez D. F., Lopez-Romero E., Zinker S., et al. (2007). Endoplasmic reticulum alpha-glycosidases of Candida albicans are required for N glycosylation, cell wall integrity, and normal host-fungus interaction. Eukaryot. Cell 6, 2184–2193. 10.1128/EC.00350-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-Montes H. M., Ponce-Noyola P., Villagómez-Castro J. C., Gow N. A. R., Flores-Carreón A., López-Romero E. (2009). Protein glycosylation in Candida. Future Microbiol. 4, 1167–1183. 10.2217/fmb.09.88 [DOI] [PubMed] [Google Scholar]
- Munro C. A., Bates S., Buurman E. T., Hughes H. B., Maccallum D. M., Bertram G., et al. (2005). Mnt1p and Mnt2p of Candida albicans are partially redundant alpha-1,2-mannosyltransferases that participate in O-linked mannosylation and are required for adhesion and virulence. J. Biol. Chem. 280, 1051–1060. 10.1074/jbc.M411413200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murad A. M., Lee P. R., Broadbent I. D., Barelle C. J., Brown A. J. (2000). CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast 16, 325–327. [DOI] [PubMed] [Google Scholar]
- Nailis H., Coenye T., Van Nieuwerburgh F., Deforce D., Nelis H. J. (2006). Development and evaluation of different normalization strategies for gene expression studies in Candida albicans biofilms by real-time PCR. BMC Mol. Biol. 7:25. 10.1186/1471-2199-7-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea M. G., Joosten L. A., van der Meer J. W., Kullberg B. J., van de Veerdonk F. L. (2015). Immune defence against Candida fungal infections. Nat. Rev. Immunol. 15, 630–642. 10.1038/nri3897 [DOI] [PubMed] [Google Scholar]
- Odani T., Shimma Y., Tanaka A., Jigami Y. (1996). Cloning and analysis of the MNN4 gene required for phosphorylation of N-linked oligosaccharides in Saccharomyces cerevisiae. Glycobiology 6, 805–810. 10.1093/glycob/6.8.805 [DOI] [PubMed] [Google Scholar]
- Orlean P. (2012). Architecture and biosynthesis of the Saccharomyces cerevisiae cell wall. Genetics 192, 775–818. 10.1534/genetics.112.144485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.-N., Song Y., Cheon S. A., Kwon O., Oh D.-B., Jigami Y., et al. (2011). Essential role of YlMPO1, a novel Yarrowia lipolytica homologue of Saccharomyces cerevisiae MNN4, in Mannosylphosphorylation of N- and O-linked glycans. Appl. Environ. Microbiol. 77, 1187–1195. 10.1128/AEM.02323-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prill S. K., Klinkert B., Timpel C., Gale C. A., Schroppel K., Ernst J. F. (2005). PMT family of Candida albicans: five protein mannosyltransferase isoforms affect growth, morphogenesis and antifungal resistance. Mol. Microbiol. 55, 546–560. 10.1111/j.1365-2958.2004.04401.x [DOI] [PubMed] [Google Scholar]
- Sanguinetti M., Posteraro B., Lass-Florl C. (2015). Antifungal drug resistance among Candida species: mechanisms and clinical impact. Mycoses 58(Suppl. 2), 2–13. 10.1111/myc.12330 [DOI] [PubMed] [Google Scholar]
- Santos E. O. L., Azzolini A. E. C. S., Lucisano-Valim Y. M. (2015). Optimization of a flow cytometric assay to evaluate the human neutrophil ability to phagocytose immune complexes via Fcγ and complement receptors. J. Pharmacol. Toxicol. Methods 72, 67–71. 10.1016/j.vascn.2014.10.005 [DOI] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539–539. 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton D. R., Masuoka J., Hazen K. C. (2005). Surface hydrophobicity changes of two Candida albicans serotype B mnn4Δ mutants. Eukaryotic Cell 4, 639–648. 10.1128/EC.4.4.639-648.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo-Esquivel E., Franco B., Flores-Martinez A., Ponce-Noyola P., Mora-Montes H. M. (2016). Purification of single-atranded cDNA based on RNA degradation treatment and adsorption chromatography. Nucleosides Nucleotides Nucleic Acids 35, 404–409. 10.1080/15257770.2016.1184277 [DOI] [PubMed] [Google Scholar]
- Vyas V. K., Barrasa M. I., Fink G. R. (2015). A Candida albicans CRISPR system permits genetic engineering of essential genes and gene families. Sci. Adv. 1:e1500248. 10.1126/sciadv.1500248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.-H., Nakayama K. I., Shimma Y. I., Tanaka A., Jigami Y. (1997). MNN6, a member of the KRE2/MNT1 family, is the gene for mannosylphosphate transfer in Saccharomyces cerevisiae. J. Biol. Chem. 272, 18117–18124. 10.1074/jbc.272.29.18117 [DOI] [PubMed] [Google Scholar]
- Wilson R. B., Davis D., Enloe B. M., Mitchell A. P. (2000). A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast 16, 65–70. [DOI] [PubMed] [Google Scholar]
- Wilson R. B., Davis D., Mitchell A. P. (1999). Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181, 1868–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.-R., Idanpaan-Heikkila I., Fischer W., Tuomanen E. I. (1999). Pneumococcal licD2 gene is involved in phosphorylcholine metabolism. Mol. Microbiol. 31, 1477–1488. 10.1046/j.1365-2958.1999.01291.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.