Fig. 4.
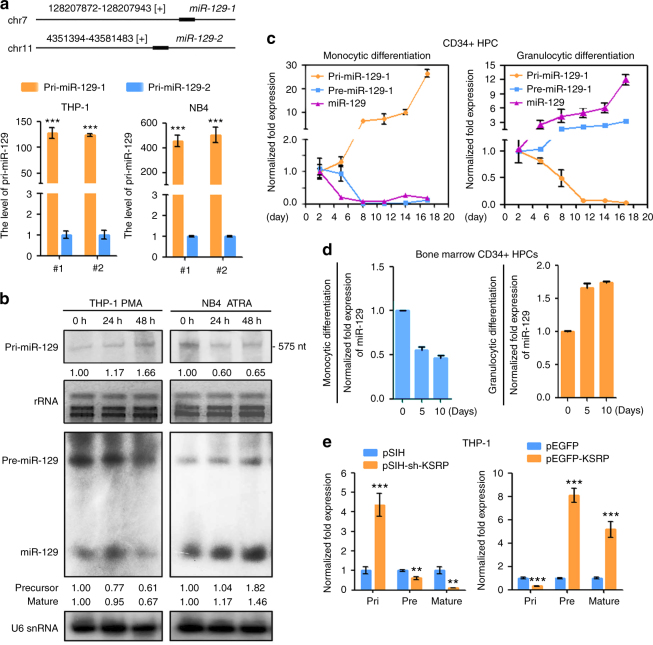
KSRP regulates primary miRNA processing in myeloid cells. a Upper: schematic representation of two miR-129 loci in the human genome; lower: qPCR analysis of the relative abundance of the two pri-129 transcripts in THP-1 and NB4 cells. The expression of each transcript was normalized to pri-129-2 using the ΔΔCq method. Three technical replicates from a single experiment representative of two independent experiments. b Northern blot analysis of miR-129 transcripts in THP-1 cells during monocyte differentiation and NB4 cells during granulocyte differentiation. The upper panels show the hybridization of pri-129-1 with digoxigenin (DIG)-labeled RNA probes; rRNA was used as a loading control. The lower panels show the hybridization of pre-miR-129-1 and miR-129 with isotope-labeled DNA probes; U6 snRNA was used as a loading control. Relative expression of pri-, pre-, and mature miR-129 was quantified using Image J software. c qPCR analysis of the expression of pri-129-1, pre-miR-129-1 and miR-129 in the CD34+ HPC models of monocytic and granulocytic differentiation. d qPCR analysis of the expression of miR-129 in the monocytic and granulocytic differentiation of bone marrow derived CD34+ HPCs. e Left: qPCR of the expression of pri-129-1, pre-miR-129-1 and miR-129 in THP-1 cells transfected with pSIH-sh-KSRP or pSIH control. Right: qPCR of the expression of pri-129-1, pre-miR-129-1 and miR-129 in THP-1 cells transfected with pEGFP-KSRP or the control. Three technical replicates from a single experiment. Data are shown as means±s.d. **P < 0.01, ***P < 0.001, Student’s t-test