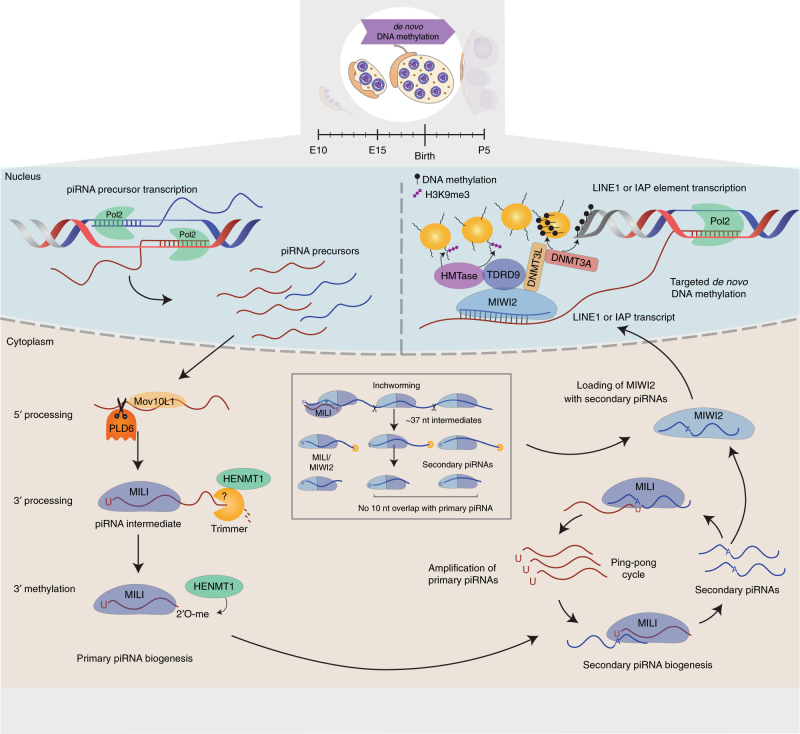
Fig. 2.
Biogenesis and function of pre-pachytene piRNAs in the mouse male germline. Pre-pachytene piRNAs are expressed during embryonic development from genomic loci that often display bidirectional transcription. piRNA precursors are exported to the cytoplasm and directed to the processing machinery. After 5′ end processing by PLD6, the piRNA intermediates associate with MILI. The 3′ ends of piRNA intermediates are then trimmed and 3′ methylated by Trimmer and HENMT1, respectively, to generate mature primary piRNAs bound to MILI. These complexes can then engage in the ping-pong cycle leading to the cleavage of complementary antisense transcripts and the production of secondary piRNAs bound to either MILI or MIWI2. Secondary piRNAs are characterized by a 10 nt overlap in their 5′ ends with primary piRNAs as a result of MILI-directed cleavage. Loading of MIWI2 with secondary piRNAs induces its translocation into the nucleus, where it functions in the transcriptional silencing of transposable elements (TEs). This involves the recruitment of the de novo DNA methylation machinery (DNMTs) as well as histone methyltransferases (HMTase) resulting in targeted DNA methylation and H3K9me3-mediated heterochromatinization of TE loci, such as LINEs and IAPs. An additional piRNA biogenesis mechanism, called inchworming, can produce MILI- and MIWI2-associated piRNAs from primary piRNA precursors without the characteristic 10 nt overlap with other piRNA species