Fig. 1.
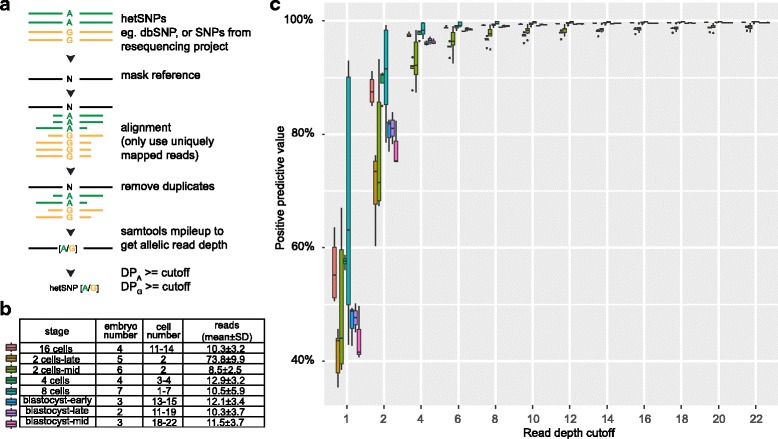
Accuracy of hetSNP calling from scRNA-seq data. a Method to compute allelic read depths at candidate SNP sites. b Summary of the mouse scRNA-seq dataset from 34 embryos at different developmental stages; each stage had multiple embryos and a different number of single cells (the minimal and maximal are provided) were analyzed for each embryo. Uniquely mapped reads in cells are presented as mean ± SD (in millions). c The percentages of hetSNPs (y-axis) called from mouse F1 embryos that were consistent with genotyping data. Boxplots are used to summarize data from individual embryos of the same developmental stages (see color legend in b). The x-axis depicts read depth cutoffs used in calling hetSNPs
