Summary
Recent analysis of genome sequences has identified individuals that are healthy despite carrying severe disease-associated mutations. A possible explanation is that these individuals carry a second genomic perturbation that can compensate for the detrimental effects of the disease allele, a phenomenon referred to as suppression. In model organisms, suppression interactions are generally divided into two classes: genomic suppressors which are secondary mutations in the genome that bypass a mutant phenotype, and dosage suppression interactions in which overexpression of a suppressor gene rescues a mutant phenotype. Here, we describe the general properties of genomic and dosage suppression, with an emphasis on the budding yeast. We propose that suppression interactions between genetic variants are likely relevant for determining the penetrance of human traits. Consequently, an understanding of suppression mechanisms may guide the discovery of protective variants in healthy individuals that carry disease alleles, which could direct the rational design of new therapeutics.
Keywords: compensatory evolution, dosage suppression, epistasis, genetic interactions, protective alleles, suppression interactions, synthetic viability
Introduction
In this era of affordable whole-genome sequencing, it remains challenging to link any of the ~4 million variants scattered across the genome of an individual to an observed phenotype. Differences in age-of-onset and severity of Mendelian disease phenotypes between family members carrying the same disease alleles are frequently observed, and can be the result of modifier genes [1–3]. Modifying mutations can either increase the severity of a disease phenotype, or they can have a protective effect and compensate for the deleterious effects of the disease mutation, a phenomenon called genetic suppression. Remarkably, in a few extreme cases, suppressor mutations could possibly be responsible for the complete rescue of the detrimental effects of genetic lesions associated with severe, early-onset Mendelian diseases [4]. However, despite these dramatic phenotypic effects, identification of suppressor mutations is challenging and requires data from either large, interrelated families or extensive genome-wide association studies (GWAS) of patients carrying the same disease allele [2, 5]. Some modifier loci, including a few potentially protective alleles, have been mapped for monogenic disorders that have a relatively high incidence, such as Huntington disease and cystic fibrosis [6,7]. However, most monogenic diseases are too rare for such systematic analyses, and GWAS generally lack the clinical information or statistical power to detect individual suppressive loci [2,8]. Although protective mutations may also affect disease severity in complex genetic diseases, such cases are likely even harder to identify, due to the additional complexity caused by variation in multiple disease-causal alleles.
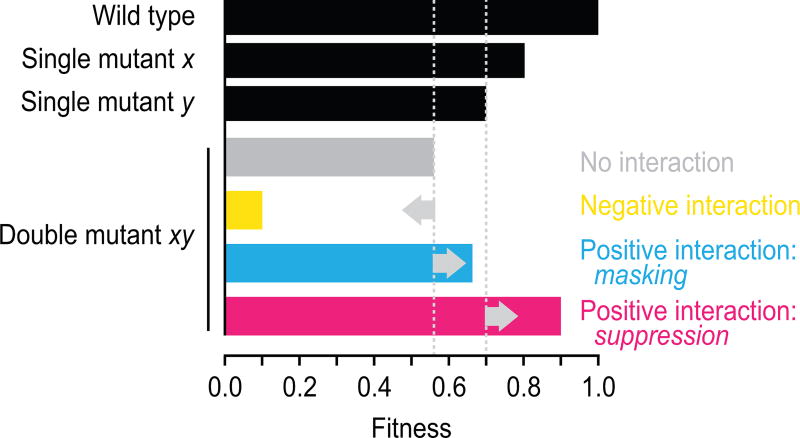
Given the challenges associated with studying suppressor mutations in outbred organisms, including humans, genetic interactions, in which two mutations combine to yield an unexpected phenotype, have mainly been studied using inbred model organisms. The use of model organisms also enables rigorous assessment of the effects of combining mutations in an otherwise isogenic background. The budding yeast Saccharomyces cerevisiae has often been the model organism of choice for these studies, due to its highly annotated genome and tractable genetics [9]. We recently completed a global network of pairwise genetic interactions between loss-of-function alleles for nearly all yeast genes [10]. The global genetic network highlighted the functional organization of a cell, revealing regulatory hubs and connecting uncharacterized genes to well-studied pathways. This study quantitatively mapped both negative interactions, which occur when the double mutant is less fit than expected based on the combined fitness of the single mutations, and positive genetic interactions, which are scored when the double mutant is more fit than expected (Fig. 1). However, suppression interactions, the most extreme type of positive interaction in which a mutant allele either partially or completely restores the fitness defect of a particular mutant (Fig. 1), were rarely detected, in part because mutant strains with a severe fitness defect do not make it through the synthetic genetic array (SGA) screening procedure.
Figure 1.
Genetic interaction classes. When two single mutants (x and y) have a relative fitness of 0.8 and 0.7, the expected fitness of the resultant double mutant (xy) based on a multiplicative model is 0.8 × 0.7 = 0.56. A negative genetic interaction, such as synthetic lethality, occurs when the observed double mutant fitness is lower than this expected fitness. A masking positive interaction occurs when the fitness of the double mutant is greater than expected, but lower or equal to that of the slowest growing single mutant. Suppression positive interactions occur when the double mutant fitness is greater than that of the slowest growing single mutant.
In model organisms, suppressors of a mutant phenotype can be identified by directly testing for an interaction between two engineered alleles, by isolating spontaneous or induced genomic mutations that can overcome the defect (genomic/genetic suppression), or by systematically testing plasmid libraries for genes that when overexpressed show a suppression phenotype (dosage suppression). Note that the distinction between genomic and dosage suppression largely relies on the method of isolation of the suppressor genes, and that genomic suppressor mutations can include dosage events, such as gene duplications. Single suppressor events have the potential to overcome extreme growth defects, and mutations have been isolated that can suppress the lethality of complete loss of function of an essential gene in yeast [11, 12]. Suppression interactions tend to occur between genes that have a close functional connection, and suppressor screens have been widely used to identify genes involved in a variety of biological pathways in bacteria, yeast, fly, and worm [13–17]. We recently mapped both spontaneous genomic suppressors and dosage suppressors of growth defects associated with yeast mutants on a large scale [17, 18]. In contrast to the interactions identified in the global genetic interaction network [10], dosage suppression interactions presumably involve hyperactive gene function, whereas spontaneous suppressor mutations can encompass the full spectrum of loss-of-function, gain-of-function, or separation-of-function events [17]. Importantly, in both cases, the mapping of dosage or genomic suppressors identifies largely novel interactions not seen in large-scale genetic interaction studies involving either complete or partial loss-of-function (hypomorphic) alleles, and thus suppression networks enhance our understanding of the functional wiring diagram of a yeast cell [17, 18].
Suppression interactions can be intragenic, occurring between two mutations within the same gene, or extragenic, involving mutations in different genes [19,20]. Intragenic suppressors include true revertants that restore the original protein sequence, revertants to another amino acid at the same codon that restores protein function, or second site mutations that may improve the mutant protein’s stability or restore the correct reading frame after a frameshift mutation [21]. In this review, we focus on extragenic suppressors and the different ways in which they can arise. Although we focus on the effects of single suppressor genes, multiple mutations can potentially combine to yield suppression phenotypes [22]. We extend previous suppressor classifications [21,23,24] to include dosage interactions, and highlight similarities and differences between spontaneous suppressor mutations and dosage suppressors. We organize the suppression interactions into distinct mechanistic categories based on the functional relationship between the gene that is being suppressed, which we refer to as the “query” gene, and the suppressor gene. As thousands of papers have been published that describe individual suppression interactions in various model organisms, we largely focus on suppression mechanisms that are prevalent in our systematic large-scale suppression networks in yeast [17,18]. Note that we expect the prevalence of suppression classes to vary depending on the experimental conditions and system, the level of gene expression, and type of allele being suppressed, among other factors. Finally, we describe how mapping suppression interactions in model organisms may help identify mutations that are protective against disease in the genomes of resilient individuals.
Mechanisms of suppression between functionally related genes
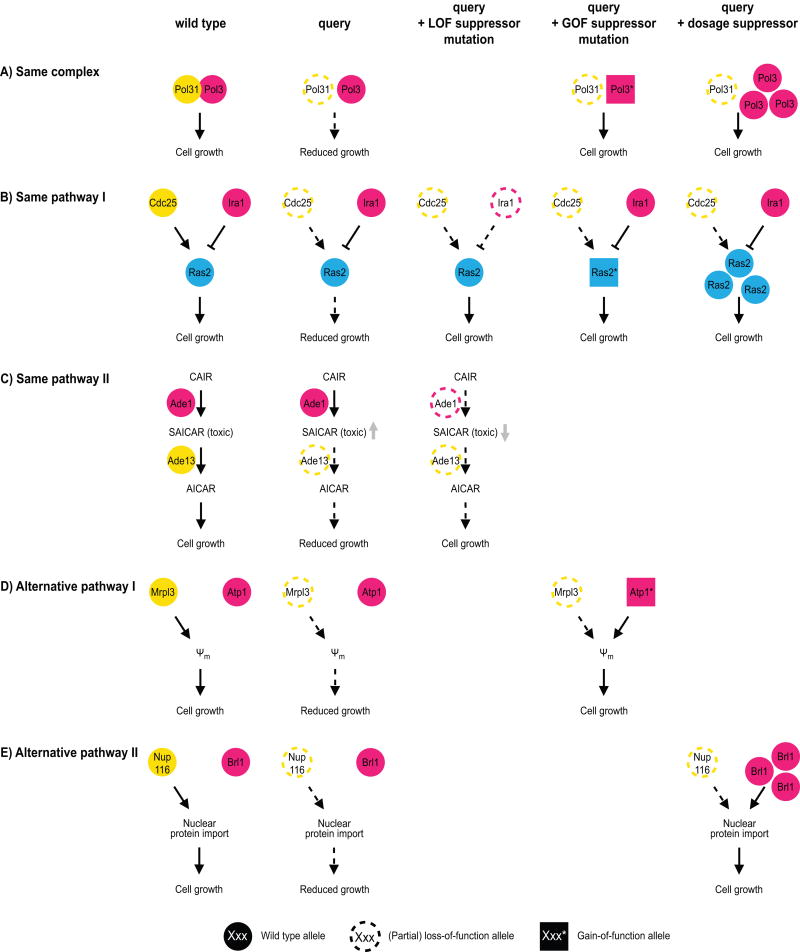
In bacteria, fungi, fly and worm, most suppression interactions reported in either small-scale or systematic studies occur between genes that are annotated to the same biological process [14, 16, 17, 25–27]. These functional connections can be further divided into three subclasses (Fig. 2). The first subclass includes interactions in which both the query and the suppressor genes encode members of the same protein complex (Fig. 2A) [17, 25]. In our large-scale suppression datasets in yeast, ~5% of genomic suppression interactions, and ~10% of dosage suppression interactions occurred between genes encoding components of the same complex (Table 1) [17, 18]. This type of interaction has been described for both prokaryotes and eukaryotes, and may represent a gain-of-function mechanism [28]. For example, partial loss-of-function mutations in DNA polymerase δ subunit Pol31 lead to a growth defect in yeast, probably due to reduced DNA replication [29]. This phenotype can be suppressed by a gain-of-function mutation in the gene encoding the DNA polymerase catalytic subunit, POL3 [17], or by overexpressing POL3 [29] (Fig. 2A). There are various possible mechanisms for “within-complex” gain-of-function suppression: 1) allele-specific suppressor mutations may compensate for a particular point-mutation in the query gene by restoring a protein-protein interaction in a “lock and key” type of model [16, 28]; 2) overexpression of a complex component may increase recruitment of a mutated query protein to the complex [30]; 3) overexpression of one paralog may compensate for the lack of the other [25]; or 4) gain-of-function mutations in a gene encoding a complex component may restore complex function in the absence of the query, by stabilizing a multimeric complex [18] or by making the function of the query subunit obsolete [31]. Finally, same-complex suppressor mutations can also be loss-of-function events, and may eliminate toxicity associated with a partially functional complex, as has been described for nicotinic acetylcholine receptor subunits in the worm [32].
Figure 2.
Mechanisms of suppression between functionally related genes. Three mechanisms of suppression between genes encoding proteins that function within the same biological process are illustrated using examples from budding yeast: suppression between members of the same complex, between members of the same pathway, or between members of alternative pathways. Query genes are colored yellow, while suppressor genes are magenta or cyan. Wild type alleles are represented as filled circles, partial or complete loss-of-function (LOF) alleles as open circles with a dashed border, and gain-of-function (GOF) alleles as filled squares. A: The query and suppressor genes encode members of the same complex. For example, either gain-of-function mutations in, or overexpression of, POL3 can restore polymerase activity in the presence of a partial loss-of-function allele of POL31. B,C: The suppressor and query are members of the same pathway. B: The genes can have opposite effects on the output of the pathway, or the suppressor can function downstream of the query protein. For instance, the growth defect caused by loss-of-function mutations in the Ras2-activating protein Cdc25 can be suppressed by mutation of Ira1, which inhibits Ras2. Suppression can also take place by increasing Ras2 activity, either by overexpression or by gain-of-function mutations in the RAS2 gene. C: The suppressor gene can function upstream of the query protein. For example, loss of Ade13 leads to a growth defect due to increased accumulation of a toxic metabolite SAICAR. This can be suppressed by loss of upstream pathway members, so that SAICAR does not get produced. D,E: The suppressor gene is part of an alternative, yet related, pathway, whose function can be slightly altered to restore the missing activity. D: The absence of the mitochondrial ribosomal protein Mrpl3 leads to a fitness defect due to a reduction in the mitochondrial membrane potential (Ψm). This can be restored by gain-of-function mutations in the ATP synthase subunit Atp1. E: An example of a dosage suppression interaction between members of alternative pathways. The fitness defect of nup116 mutants is the result of reduced nuclear protein import, which can be suppressed by overexpression of BRL1, a gene that changes the composition of the nuclear membrane.
Table 1. Relative frequencies of mechanistic suppression classes in genomic and dosage suppression datasets in yeast.
Suppression interactions were divided into distinct mechanistic classes based on the functional relationship between the query and suppressor genes, as described in [17]. The “informational” class includes suppressor mutations in tRNA genes. The “protein expression” class consists of suppressor genes encoding components of the mRNA decay pathway, ribosomal subunits, or transcription factors. The “protein stability” class involves suppressor genes encoding chaperones or proteins with a role in protein degradation. Values represent the percentage of suppression interactions assigned to a specific mechanistic category. Throughput: LT, low-throughput, refers to suppression interactions identified via the curation of biomedical literature, which mainly includes studies focused on the functional annotation of a particular gene; HT, high-throughput, indicates unbiased, large-scale analyses of suppression interactions. Query allele types: hypomorph, partial loss-of-function allele such as a temperature sensitive allele; deletion, complete loss-of-function allele; all, includes partial and complete loss-of-function alleles.
| Genomic suppression (% suppression interactions) |
Dosage suppression (% suppression interactions) |
|||||
|---|---|---|---|---|---|---|
| throughput | LT | HT | HT | LT | HT | HT |
| query allele-type | all | hypomorph | deletion | hypomorph | hypomorph | all |
| reference | [17] | [17] | [17] | [18] | [18] | [33] |
|
| ||||||
| Functional mechanisms | 52.7 | 19.4 | 61.3 | 65.0 | 45.9 | 23.6 |
| Same complex | 6.9 | 5.3 | 5.1 | 19.3 | 10.8 | 6.5 |
| Same pathway | 10.5 | 1.8 | 5.9 | 7.7 | 1.6 | 4.2 |
| Alternative pathway | 7.8 | 3.5 | 29.9 | 5.4 | 0.5 | 4.2 |
| Unknown functional connection | 27.5 | 8.8 | 20.4 | 32.6 | 33.0 | 8.7 |
|
| ||||||
| General mechanisms | 11.0 | 49.0 | 7.3 | 9.5 | 26.3 | 16.4 |
| Informational | 0 | 1.7 | 0 | 0 | 0 | 0 |
| Protein expression | 7.0 | 24.5 | 4.4 | 6.3 | 22.7 | 14.7 |
| Protein stability | 4.0 | 22.8 | 2.9 | 3.2 | 3.6 | 1.7 |
|
| ||||||
| Unknown mechanism | 36.3 | 31.6 | 31.4 | 25.5 | 27.8 | 60.0 |
In a second subclass of suppression interactions between functionally related genes, a query mutant phenotype is suppressed by a mutation in a gene that is annotated to the same pathway but does not encode a member of the same complex (Fig. 2B,C). This mechanistic class is equally prevalent in dosage and genomic suppression datasets, and explains ~2–6% of all unbiased suppression interactions mapped in yeast (Table 1) [17, 18, 33]. In the case of loss-of-function suppressor mutations, the suppressor gene often has antagonistic effects compared to the query gene. For instance, in yeast, the temperature-sensitive growth defect caused by mutations in the gene encoding the guanine nucleotide exchange factor Cdc25 can be suppressed by a loss-of-function mutation in the gene encoding the cognate GTPase activating protein Ira1 (Fig. 2B) [17]. Alternatively, the requirement for Cdc25 can be suppressed by activation of the downstream effector, the GTPase Ras2, through either RAS2 overexpression of by gain-of-function mutations in RAS2 [34, 35]. Specific gain- or separation-of-function suppressor mutations can also affect interactions of the suppressor protein with either DNA or another protein, and change the dynamics of an interaction that would normally be regulated by the query protein [31, 36]. Alternatively, suppression can occur by loss-of-function mutations in downstream pathway members when the upstream query protein negatively regulates the downstream suppressor gene [37]. For example, the embryonic lethality caused by loss of Mdm2, which encodes a negative regulator of the cell cycle arrest-inducing protein p53, can be suppressed by loss of p53 in mice [38, 39]. In rare cases, the suppressor mutations may mimic the effect of a posttranslational modification normally generated by the query protein, such as an acetylation-mimicking suppressor mutation in cohesion subunit Smc3, which compensates for the fitness defect caused by reduced expression of acetyltransferase Eco1 in yeast [40]. Finally, loss-of-function mutations in upstream members of a pathway can also cause suppression. For example, the essential yeast gene ADE13 encodes an enzyme in the 10-step de novo purine synthesis pathway that converts 5-phosphoribosylpyrophosphate into inosine monophosphate, from which adenine and guanine are formed. Unbiased suppression analysis in yeast showed that loss of any of the genes upstream of ADE13 in the pathway can rescue the lethality caused by loss of ADE13, while deletion of downstream genes could not (Fig. 2C) [11]. None of the other genes in the pathway are essential, suggesting that loss of Ade13 leads to the accumulation of a toxic intermediate, in this case (S)-2-[5-Amino-1-(5-phospho-D-ribosyl)imidazole-4-carboxamido]succinate or SAICAR, which can be prevented by inactivation of any of the upstream pathway members. Although suppression analysis can often be used to deduce the order of action of pathway components, these examples illustrate that inference about pathway structure depends on the pathway, the type of regulation, and the nature of the mutation, and additional molecular methods are often required to reliably determine gene order [41, 42].
The third mechanistic class of suppression by functionally related genes, involves suppression by mutation of members of a different, but related, pathway (Fig. 2D,E). In this scenario the phenotype caused by the absence of a specific cellular function is suppressed when an alternative pathway is rewired to compensate for the missing activity. This mechanism of suppression is observed for <10% of all suppression interactions in yeast (Table 1) [17, 18]. However, this type of suppression is over-represented among genomic suppressors of deletion alleles in our unbiased yeast dataset (30% of interactions, Table 1), reflecting many instances of suppression of mitochondrial transcription or translation mutants that exhibit a growth defect due to decreased protein import into the mitochondria. In these cases, growth phenotypes can be suppressed by specific mutations in the mitochondrial ATP synthase that restore mitochondrial protein import by reversing the activity of the mitochondrial ATP synthase, such that it generates ADP3− instead of ATP4− [17] (Fig. 2D). The charge difference between these two nucleotide phosphates can be exploited by adenine nucleotide translocators to rebuild the mitochondrial membrane potential, which is lost in the absence of mitochondrial transcription or translation mutants, and is thought to be required for protein import into the mitochondria [43]. In an example of a dosage suppression interaction between alternative pathway members, loss of specific members of the nuclear pore complex in yeast leads to a growth defect due to reduced protein import into the nucleus (Fig. 2E) [12]. Both nuclear protein import and fitness of these mutants can be restored by overexpression of nuclear envelope protein Brl1, probably by reducing nuclear membrane fluidity [12].
Around 10–30% of all suppression interactions occur between genes that function in the same biological process but that lack a pathway or complex annotation for at least one of the interacting genes (Table 1). Improved annotation of complex or pathway membership will aid further quantification of mechanisms of suppression between functionally related genes.
General mechanisms of suppression
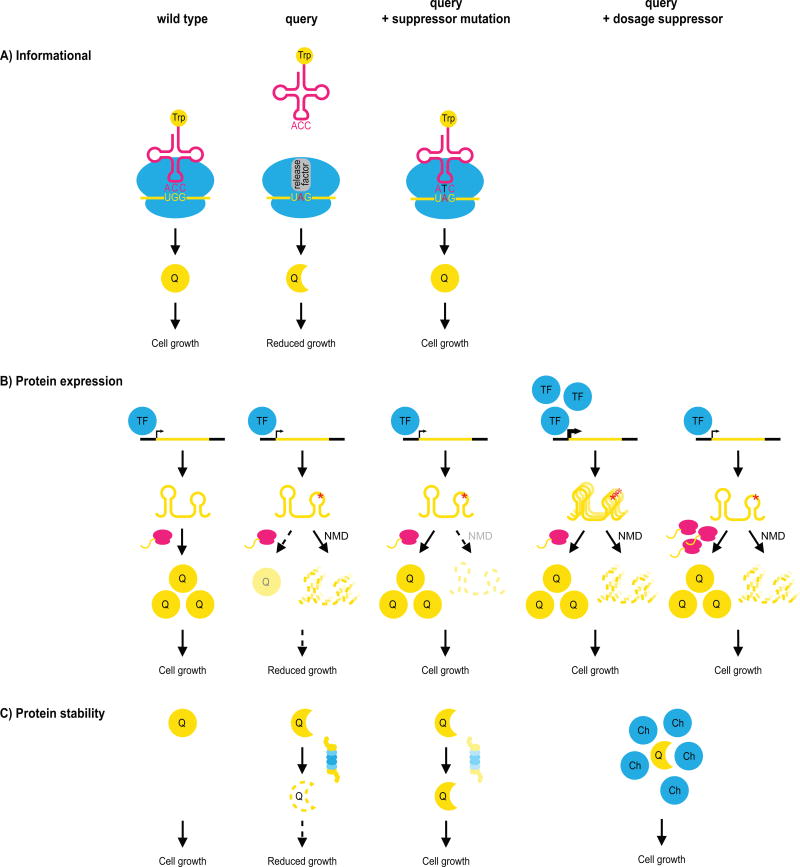
In addition to suppression interactions between genes that share a functional connection, suppression interactions involving partial loss-of-function alleles, or hypomorphs, reveal three different and more general classes of suppressors that affect the translation of the query mutation, the expression of the query gene, or the stability of its gene-product (Fig. 3, Table 1). These suppressors are often allele-specific, and can be either dosage or genomic suppressors, although the actual suppressor genes and mechanistic details slightly differ [17, 18].
Figure 3.
General mechanisms of suppression. General suppression interactions among pairs of genes that do not share a close functional relationship are illustrated. Often, general suppression is associated with partial loss-of-function query alleles that carry mutations that destabilize the protein or mRNA, leading to a fitness defect caused by reduced levels of the query protein (‘Q’). A: A suppressor mutation may occur in the translational machinery, such that the genetic code is changed, and the query mutation is reinterpreted. The example illustrates suppression caused by mutation of the anticodon of a tRNA to make it recognize a premature stop codon in the query allele, and introduce an amino acid in its place. B: Partial loss-of-function query alleles can also be suppressed by increasing protein expression, for instance through decreased degradation of the mutant mRNA via mutation of the nonsense-mediated mRNA decay (NMD) pathway, or via increased transcription or translation of the query gene or mRNA. TF: transcription factor. C: Partial loss-of-function mutations can be suppressed by loss of a member of the protein degradation pathway or by overexpressing a chaperone protein (‘Ch’). Both of these mechanisms may expand the pool of partially functional query protein.
The first class of general suppressors are the informational suppressors. Informational suppression refers to the alteration of the translational machinery, enabling a different interpretation of the query mutation [20, 21]. Informational suppression often occurs by modifying the genetic code through mutations in the anticodon of tRNA molecules. For example, a premature stop codon may be recognized by a mutated tryptophan-carrying tRNA, such that tryptophan is now introduced at the site of the stop codon, allowing for continued translation of the mutant transcript (Fig. 3A). In yeast, informational suppression can be greatly enhanced by switching the confirmation of translation release factor Sup35 to the [PSI+] prion state, which can also function as a weak suppressor of premature stop codons on its own [44]. In [PSI+] cells, most Sup35 protein molecules are part of amyloid aggregates and thus unavailable for translation termination, leading to increased read-through at premature stop codons [45]. Informational suppression has been described in literature for many years (see for example [46–48]), but is rarely reported in recent high-throughput experiments (incidence <2%, Table 1) [17]. This is likely due to the development of genome-wide deletion mutant collections for most popular model organisms [49–51], making mutant alleles carrying premature stop codons, and thus this type of suppression, virtually obsolete in systematic suppression screens.
A suppressor can also compensate for the growth defect of a particular query by increasing the expression of the query mutant protein (Fig. 3B). This type of suppression can be achieved in three ways: 1) by stabilizing a mutant mRNA by perturbing pathways or complexes that regulate mRNA decay; 2) by increasing transcription or translation of the mutant gene or mRNA; or 3) with compensating mutations that alter splice-site recognition or promote alternative splicing. Spontaneous suppressor mutations in yeast frequently fall into the first category (~25% of genomic suppressors of partial loss-of-function alleles, Table 1), and are often loss-of-function mutations in NMD2 or NAM7, which encode members of the nonsense-mediated mRNA decay pathway [17]. These mutations do not solely suppress query alleles carrying premature stop codons, but also alleles carrying other point-mutations or in which the 3’UTR has been disrupted (decreased abundance by mRNA perturbation, DAmP, alleles [52]). Dosage suppression interactions generally fall into the second category, and affect transcription or translation (Fig. 3B). For example, increased transcription can result from overexpression of a transcription factor that activates the query gene [18]. The mechanisms by which increased protein synthesis could cause suppression are less obvious. Genes encoding ribosomal subunits are often found as dosage suppressors [18, 33, 53], but it seems unlikely that overexpression of a single subunit of the ribosome could lead to increased translation. In apparent contrast, partial knockdown of ribosomal subunits can rescue a variety of mutant phenotypes in the worm Caenorhabditis elegans [27, 54]. However, a recent study showed a correlation between ribosome composition and fitness, suggesting that ribosomal subunit stoichiometry may either regulate protein synthesis, or affect cell cycle progression [55]. Other studies suggest that some individual ribosomal subunits may possess extraribosomal functions, and can for example function as chaperones that increase the stability of certain proteins [53, 56]. Thus, although the exact nature of suppression by altered expression of ribosomal subunits remains unclear, it may be related to modified expression of either the query protein or of another protein that can compensate for the query protein defects. Together, these mechanisms of increased transcription and translation explain ~20% of dosage suppression interactions in high-throughput yeast datasets (Table 1) [18]. The third category, suppression by mutations in the splicing machinery that lead to altered splicing, is rarely described in yeast, because only a small fraction of S. cerevisiae genes have introns, but is prevalent in organisms with extensive splicing, such as the worm [24].
Finally, suppression of partial loss-of-function alleles can occur by affecting protein stability or degradation (Fig. 3C). In our large-scale study of genomic suppressors in yeast, 23% of the partial loss-of-function alleles were suppressed by this mechanism (Table 1), and mainly carried loss-of-function mutations in the gene encoding the ubiquitin-protein ligase San1 which targets proteins with exposed hydrophobic residues for proteasome-dependent degradation [17]. Partial inactivation of pathways that promote protein turnover may increase the levels of the mutant query protein, thereby compensating for the phenotype caused by decreased protein availability or activity (Fig. 3C). Similarly, dosage suppression can occasionally (2–4% of all interactions) be achieved by overexpression of a chaperone that stabilizes the query mutant protein, and thus enlarges the pool of active query protein (Fig. 3C, Table 1) [18, 33].
General suppression mechanisms tend to be rarely described in studies focused on the functional annotation of a particular gene (Table 1), but are highly prevalent in systematic datasets [17, 18, 33], and explain nearly 50% of the genomic suppressors and 26% of dosage suppressors of partial loss-of-function alleles (Table 1) [17]. An additional general class of suppressors has been identified in worm, involving mutations that weakly suppress several unrelated mutant phenotypes, probably by changing the general physiology of the animal [24]. Around 30% of all suppression interactions do not fall into any of the above-mentioned functional or general categories (Table 1), partially because the functional annotation of the yeast genome is incomplete. As both the genomic and dosage suppression networks are far from complete, additional mechanisms of suppression may emerge upon expansion of these networks.
Evidence for suppression interactions among human genetic variants
Most suppression interactions have been identified in model organisms, and the question remains as to whether these interactions play a significant role in determining human genetic traits. A typical human genome carries ~4 million variants, including 85 heterozygous and 35 homozygous null alleles [57], and 74 amplifications [58]. As human variants of interest can be either single nucleotide polymorphisms or gene duplication events, we will not make the distinction between genomic and dosage suppressors, but instead consider human genetic suppression in general. As discussed above, suppression is widespread in haploid microorganisms. A quantitative analysis uncovered suppressors for >65% of the tested deletion alleles in yeast [25], and this number is likely even higher for partial loss-of-function mutants. Arguably though, the frequency of these interactions may differ in naturally diploid populations in which mutations in one copy of the gene may be buffered by the second copy. However, several points of evidence suggest that suppression is likely prevalent in higher organisms as well. First, fitter mutants can be readily isolated in more complex model organisms, such as mice. For instance, by screening over a thousand mutagenized mice, dominant suppressor mutations were identified for mutations in mouse orthologues of genes associated with the human disorders thrombocytopenia (a lack of blood platelets) and Rett syndrome [59, 60]. Second, the degree of severity of the phenotype caused by a specific mutation can differ between individuals of the same species, and between different species. For example, ~6% of essential yeast genes were found to be uniquely essential in only one of two closely related yeast strains, ~1% of pathogenic variants in Drosophila melanogaster are fixed in other flies, and around 6% of human disease-causing variants are found in orthologous genes in mammals, and seem to be the result of one rescuing mutation [61–63]. These observations suggest that suppression interactions may be common among human genetic variants.
Some specific examples of suppression interactions affecting disease severity have been described. In a few extreme cases, individuals have been reported in whom a Mendelian disease phenotype was completely suppressed [4, 64]. For example, most individuals that are homozygous for the recessive nonsyndromic deafness locus DFNB26 have hearing loss, but several members of one family have been identified who have normal hearing despite carrying two defective copies of DFNB26 [64]. A dominant suppressor locus, DFNM1, was identified, which segregates through the family and protects family members carrying it against hearing loss [64]. In a similar example in dogs, golden retrievers were identified that only showed mild symptoms of muscular dystrophy despite lacking the dystrophin gene, a mutation that is normally causal of the disease [65]. The suppression phenotype was mapped to a mutation in a transcription factor binding site that leads to increased expression of Jagged1, suggesting that this gene may represent a therapeutic target [65]. Suppression interactions are also of key importance in understanding how genetic differences can affect complex disease phenotypes, such as cancer progression. In cancer, suppression interactions that make cells grow faster are associated with a poorer prognosis. Suppressor studies in cultured cells revealed that the growth defect caused by absence of the tumor suppressor gene BRCA1 can be suppressed by loss of either 53BP1 or REV7 (MAD2L2) [66, 67]. BRCA1 has a role in the repair of DNA double-strand breaks by homologous recombination (HR), and loss of BRCA1 leads to toxic chromosomal deletions or rearrangements due to increased repair of double-strand DNA breaks by nonhomologous end joining. 53BP1 and REV7 both negatively regulate HR initiation, and loss of one of these proteins can suppress BRCA1-mutant phenotypes by allowing double-strand break repair to occur by the compromised, but more accurate, HR machinery [68]. These findings partially translate to clinical phenotypes, as loss of 53BP1 is more frequent in tumors with BRCA1 mutations, and is associated with aggressive tumors and low survival rates [66]. In addition, although BRCA1-deficiency makes tumor cells uniquely sensitive to inhibitors of poly(ADP-ribose) polymerases (PARPs), subsequent 53BP1 loss abrogates this hypersensitivity and confers resistance to PARP-inhibitors [69]. The occurrence of suppressing mutations can thus have a major impact on both tumor malignancy and drug response.
To summarize, based on the ease of suppression isolation in model organisms, the frequency of suppression in natural populations, and the few known examples of suppression in determining disease severity, we conclude that genetic suppression is likely an important determinant of human traits.
Applying suppression to identify modifiers of human disease
How has the mapping of suppression interactions in model organisms contributed to understanding suppression in humans? There is evidence that the suppression mechanisms we have identified in yeast are applicable to other organisms as well. Although not studied systematically, similar mechanisms are generally found in suppression studies in fly and worm [15, 24, 27]. Importantly, the mechanisms that were identified as common in yeast, can be used to decipher human genotype-to-phenotype relationships. For example, one general mechanism of suppression that was widespread in our suppression studies, is the compensation of the fitness defects of partial loss-of-function mutants by decreased mRNA decay [17]. Many diseases arise as a consequence of a nonsense or frameshift mutation that introduces a premature termination codon [70]. In such cases, nonsense mediated mRNA decay (NMD) can be either protective, by removing a transcript that would otherwise result in a toxic protein, or detrimental, because degradation of the mutated mRNA prevents translation and accumulation of truncated peptides that retain residual activity [71, 72]. It thus seems likely that inter-individual variability in NMD efficiency leads to differences in disease presentation, and reduced NMD may be protective of specific disease-variants. In fact, in certain genetic disorders, such as Duchenne muscular dystrophy, treatment options are focused on increasing translation of the disease-associated mutant mRNA [73]. Some of the other above-mentioned general mechanisms of suppression, such as those affecting protein stability, may have similar relevance to human disease.
In addition to their use in uncovering general characteristics of suppression interactions, screens in model organisms for mutations that can overcome phenotypes caused by human disease alleles may directly identify novel targets for drug discovery. This especially holds true in cases where the suppressor mutations are loss-of-function events, as most small molecules are thought to inhibit the function of the protein they bind. In a screen with mice, detrimental phenotypes triggered by mutation of Mecp2, which causes the autism spectrum disorder Rett syndrome in human, were found to be suppressed by mutation of Sqle, which encodes an enzyme involved in cholesterol synthesis [60]. Administration of statins that repress cholesterol synthesis, improved the fitness of Mecp2 mutant mice, showing the potential of suppression screens to identify new treatment possibilities [60].
Conclusions and prospects
Suppression analysis is often applied to dissect biological pathways and functional relationships. In this review, we have summarized the various ways in which genomic and dosage suppression can occur, and the complementary information that can be obtained with different types of suppression screens. Our recent large scale analyses of suppression in yeast allowed us to estimate the prevalence of these mechanistic suppression classes [17, 18]. We anticipate that a thorough understanding of mechanisms of suppression and their frequency could be exploited to decipher genotype-to-phenotype relationships, and may help narrow down the search for protective mutations amongst the millions of variants in resilient individuals that carry a disease-causing mutation but do not show a disease phenotype. Also for more complex traits, insight into the molecular mechanisms by which suppression can occur may shed light on how genetic variants interact to produce observed disease phenotypes. As one of the few known examples of suppression in humans, the interaction between BRCA1 and 53BP1 highlights how the occurrence of suppressing mutations can have a major impact on both tumor malignancy and drug response. We expect similar interactions between variants to influence the severity of disease phenotypes in other complex diseases.
Acknowledgments
We thank H. Friesen for critical comments. Functional genomics work in the Boone and Andrews labs is supported primarily by a grant from the NIH (R01HG00583) and Foundation Grants from the Canadian Institutes for Health Research (CIHR: FDN-143264; FDN-143265). J.v.L. was supported by a postdoctoral fellowship from the CIHR. C.B. and B.A. are Senior Fellows and co-Director (C.B.) of the Canadian Institute for Advanced Research Genetic Networks program. C.P. is supported by a Juan de la Cierva fellowship.
References
- 1.Nadeau JH. Modifier genes in mice and humans. Nat Rev Genet. 2001;2:165–74. doi: 10.1038/35056009. [DOI] [PubMed] [Google Scholar]
- 2.Genin E, Feingold J, Clerget-Darpoux F. Identifying modifier genes of monogenic disease: strategies and difficulties. Hum Genet. 2008;124:357–68. doi: 10.1007/s00439-008-0560-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harper AR, Nayee S, Topol EJ. Protective alleles and modifier variants in human health and disease. Nat Rev Genet. 2015;16:689–701. doi: 10.1038/nrg4017. [DOI] [PubMed] [Google Scholar]
- 4.Chen R, Shi L, Hakenberg J, Naughton B, et al. Analysis of 589,306 genomes identifies individuals resilient to severe Mendelian childhood diseases. Nat Biotechnol. 2016;34:531–8. doi: 10.1038/nbt.3514. [DOI] [PubMed] [Google Scholar]
- 5.Wilcox E, Riazuddin S, Riazuddin S. Some deafness-causing mutations can be silenced with the appropriate gene partner. Scientific World Journal. 2001;1:202–3. doi: 10.1100/tsw.2001.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guillot L, Beucher J, Tabary O, Le Rouzic P, et al. Lung disease modifier genes in cystic fibrosis. Int J Biochem Cell Biol. 2014;52:83–93. doi: 10.1016/j.biocel.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 7.Arning L. The search for modifier genes in Huntington disease - Multifactorial aspects of a monogenic disorder. Mol Cell Probes. 2016;30:404–9. doi: 10.1016/j.mcp.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 8.Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: Genetic interactions create phantom heritability. Proc Natl Acad Sci USA. 2012;109:1193–8. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Botstein D, Fink GR. Yeast: an experimental organism for 21st century biology. Genetics. 2011;189:695–704. doi: 10.1534/genetics.111.130765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costanzo M, VanderSluis B, Koch EN, Baryshnikova A, et al. A global genetic interaction network maps a wiring diagram of cellular function. Science. 2016;353:1381. doi: 10.1126/science.aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen P, Wang D, Chen H, Zhou Z, et al. The nonessentiality of essential genes in yeast provides therapeutic insights into a human disease. Genome Res. 2016;26:1355–62. doi: 10.1101/gr.205955.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu G, Yong MY, Yurieva M, Srinivasan KG, et al. Gene essentiality is a quantitative property linked to cellular evolvability. Cell. 2015;163:1388–99. doi: 10.1016/j.cell.2015.10.069. [DOI] [PubMed] [Google Scholar]
- 13.Forsburg SL. The art and design of genetic screens: yeast. Nat Rev Genet. 2001;2:659–68. doi: 10.1038/35088500. [DOI] [PubMed] [Google Scholar]
- 14.Jorgensen EM, Mango SE. The art and design of genetic screens: Caenorhabditis elegans. Nat Rev Genet. 2002;3:356–69. doi: 10.1038/nrg794. [DOI] [PubMed] [Google Scholar]
- 15.St Johnston D. The art and design of genetic screens: Drosophila melanogaster. Nat Rev Genet. 2002;3:176–88. doi: 10.1038/nrg751. [DOI] [PubMed] [Google Scholar]
- 16.Manson MD. Allele-specific suppression as a tool to study protein-protein interactions in bacteria. Methods. 2000;20:18–34. doi: 10.1006/meth.1999.0902. [DOI] [PubMed] [Google Scholar]
- 17.Van Leeuwen J, Pons C, Mellor JC, Yamaguchi TN, et al. Exploring genetic suppression interactions on a global scale. Science. 2016;354:599. doi: 10.1126/science.aag0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magtanong L, Ho CH, Barker SL, Jiao W, et al. Dosage suppression genetic interaction networks enhance functional wiring diagrams of the cell. Nat Biotechnol. 2011;29:505–11. doi: 10.1038/nbt.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehner B. Molecular mechanisms of epistasis within and between genes. Trends Genet. 2011;27:323–31. doi: 10.1016/j.tig.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 20.Botstein D. Decoding the language of genetics Cold Spring Harbor. New York: Cold Spring Harbor Laboratory Press; 2015. [Google Scholar]
- 21.Hartman PE, Roth JR. Mechanisms of suppression. Adv Genet. 1973;17:1–105. doi: 10.1016/s0065-2660(08)60170-4. [DOI] [PubMed] [Google Scholar]
- 22.Taylor MB, Ehrenreich IM. Higher-order genetic interactions and their contribution to complex traits. Trends Genet. 2015;31:34–40. doi: 10.1016/j.tig.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prelich G. Suppression mechanisms: themes from variations. Trends Genet. 1999;15:261–6. doi: 10.1016/s0168-9525(99)01749-7. [DOI] [PubMed] [Google Scholar]
- 24.Hodgkin J. Genetic suppression. Worm Book. 2005:1–13. doi: 10.1895/wormbook.1.59.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szamecz B, Boross G, Kalapis D, Kovacs K, et al. The genomic landscape of compensatory evolution. PLoS Biol. 2014;12:e1001935. doi: 10.1371/journal.pbio.1001935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harcombe WR, Springman R, Bull JJ. Compensatory evolution for a gene deletion is not limited to its immediate functional network. BMC Evol Biol. 2009;9:106. doi: 10.1186/1471-2148-9-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fievet BT, Rodriguez J, Naganathan S, Lee C, et al. Systematic genetic interaction screens uncover cell polarity regulators and functional redundancy. Nat Cell Biol. 2013;15:103–12. doi: 10.1038/ncb2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sujatha S, Chatterji D. Understanding protein–protein interactions by genetic suppression. J Genet. 2000;79:125–9. [Google Scholar]
- 29.Hashimoto K, Nakashima N, Ohara T, Maki S, et al. The second subunit of DNA polymerase III (delta) is encoded by the HYS2 gene in Saccharomyces cerevisiae. Nucleic Acids Res. 1998;26:477–85. doi: 10.1093/nar/26.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prelich G. Gene overexpression: uses, mechanisms, and interpretation. Genetics. 2012;190:841–54. doi: 10.1534/genetics.111.136911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Menne TF, Goyenechea B, Sanchez-Puig N, Wong CC, et al. The Shwachman-Bodian-Diamond syndrome protein mediates translational activation of ribosomes in yeast. Nat Genet. 2007;39:486–95. doi: 10.1038/ng1994. [DOI] [PubMed] [Google Scholar]
- 32.Treinin M, Gillo B, Liebman L, Chalfie M. Two functionally dependent acetylcholine subunits are encoded in a single Caenorhabditis elegans operon. Proc Natl Acad Sci USA. 1998;95:15492–5. doi: 10.1073/pnas.95.26.15492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patra B, Kon Y, Yadav G, Sevold AW, et al. A genome wide dosage suppressor network reveals genomic robustness. Nucleic Acids Res. 2016;45:255–70. doi: 10.1093/nar/gkw1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park W, Mosteller RD, Broek D. Identification of a dominant-negative mutation in the yeast CDC25 guanine nucleotide exchange factor for Ras. Oncogene. 1997;14:831–6. doi: 10.1038/sj.onc.1200893. [DOI] [PubMed] [Google Scholar]
- 35.Camonis JH, Jacquet M. A new RAS mutation that suppresses the CDC25 gene requirement for growth of Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:2980–3. doi: 10.1128/mcb.8.7.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Puddu F, Oelschlaegel T, Guerini I, Geisler NJ, et al. Synthetic viability genomic screening defines Sae2 function in DNA repair. EMBO J. 2015;34:1509–22. doi: 10.15252/embj.201590973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2:329–40. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- 38.Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–8. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 39.Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–6. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 40.Rolef Ben-Shahar T, Heeger S, Lehane C, East P, et al. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–6. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- 41.Avery L, Wasserman S. Ordering gene function: the interpretation of epistasis in regulatory hierarchies. Trends Genet. 1992;8:312–6. doi: 10.1016/0168-9525(92)90263-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stevenson BJ, Rhodes N, Errede B, Sprague GF., Jr Constitutive mutants of the protein kinase STE11 activate the yeast pheromone response pathway in the absence of the G protein. Genes Dev. 1992;6:1293–304. doi: 10.1101/gad.6.7.1293. [DOI] [PubMed] [Google Scholar]
- 43.Clark-Walker GD. Kinetic properties of F1-ATPase influence the ability of yeasts to grow in anoxia or absence of mtDNA. Mitochondrion. 2003;2:257–65. doi: 10.1016/S1567-7249(02)00107-1. [DOI] [PubMed] [Google Scholar]
- 44.Liebman SW, Sherman F. Extrachromosomal psi+ determinant suppresses nonsense mutations in yeast. J Bacteriol. 1979;139:1068–71. doi: 10.1128/jb.139.3.1068-1071.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salnikova AB, Kryndushkin DS, Smirnov VN, Kushnirov VV, et al. Nonsense suppression in yeast cells overproducing Sup35 (eRF3) is caused by its non-heritable amyloids. J Biol Chem. 2005;280:8808–12. doi: 10.1074/jbc.M410150200. [DOI] [PubMed] [Google Scholar]
- 46.Glass RE, Nene V, Hunter MG. Informational suppression as a tool for the investigation of gene structure and function. Biochem J. 1982;203:1–13. doi: 10.1042/bj2030001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gorini L. Informational suppression. Annu Rev Genet. 1970;4:107–34. doi: 10.1146/annurev.ge.04.120170.000543. [DOI] [PubMed] [Google Scholar]
- 48.Boone C, Clark KL, Sprague GF., Jr Identification of a tRNA(Gln) ochre suppressor in Saccharomyces cerevisiae. Nucleic Acids Res. 1992;20:4661. doi: 10.1093/nar/20.17.4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Giaever G, Chu AM, Ni L, Connelly C, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–91. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 50.Kim DU, Hayles J, Kim D, Wood V, et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2010;28:617–23. doi: 10.1038/nbt.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baba T, Ara T, Hasegawa M, Takai Y, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006. doi: 10.1038/msb4100050. 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schuldiner M, Collins SR, Thompson NJ, Denic V, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–19. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 53.Kabir MA, Sherman F. Overexpressed ribosomal proteins suppress defective chaperonins in Saccharomyces cerevisiae. FEMS Yeast Res. 2008;8:1236–44. doi: 10.1111/j.1567-1364.2008.00425.x. [DOI] [PubMed] [Google Scholar]
- 54.O'Rourke SM, Dorfman MD, Carter JC, Bowerman B. Dynein modifiers in C. elegans: light chains suppress conditional heavy chain mutants. PLoS Genet. 2007;3:e128. doi: 10.1371/journal.pgen.0030128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Slavov N, Semrau S, Airoldi E, Budnik B, et al. Differential stoichiometry among core ribosomal proteins. Cell Rep. 2015;13:865–73. doi: 10.1016/j.celrep.2015.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol Cell. 2009;34:3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lek M, Karczewski KJ, Minikel EV, Samocha KE, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zarrei M, MacDonald JR, Merico D, Scherer SW. A copy number variation map of the human genome. Nat Rev Genet. 2015;16:172–83. doi: 10.1038/nrg3871. [DOI] [PubMed] [Google Scholar]
- 59.Carpinelli MR, Hilton DJ, Metcalf D, Antonchuk JL, et al. Suppressor screen in Mpl−/− mice: c-Myb mutation causes supraphysiological production of platelets in the absence of thrombopoietin signaling. Proc Natl Acad Sci USA. 2004;101:6553–8. doi: 10.1073/pnas.0401496101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Buchovecky CM, Turley SD, Brown HM, Kyle SM, et al. A suppressor screen in Mecp2 mutant mice implicates cholesterol metabolism in Rett syndrome. Nat Genet. 2013;45:1013–20. doi: 10.1038/ng.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dowell RD, Ryan O, Jansen A, Cheung D, et al. Genotype to phenotype: a complex problem. Science. 2010;328:469. doi: 10.1126/science.1189015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kulathinal RJ, Bettencourt BR, Hartl DL. Compensated deleterious mutations in insect genomes. Science. 2004;306:1553–4. doi: 10.1126/science.1100522. [DOI] [PubMed] [Google Scholar]
- 63.Jordan DM, Frangakis SG, Golzio C, Cassa CA, et al. Identification of cis-suppression of human disease mutations by comparative genomics. Nature. 2015;524:225–9. doi: 10.1038/nature14497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Riazuddin S, Castelein CM, Ahmed ZM, Lalwani AK, et al. Dominant modifier DFNM1 suppresses recessive deafness DFNB26. Nat Genet. 2000;26:431–4. doi: 10.1038/82558. [DOI] [PubMed] [Google Scholar]
- 65.Vieira NM, Elvers I, Alexander MS, Moreira YB, et al. Jagged 1 Rescues the Duchenne Muscular Dystrophy Phenotype. Cell. 2015;163:1204–13. doi: 10.1016/j.cell.2015.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bouwman P, Aly A, Escandell JM, Pieterse M, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol. 2010;17:688–95. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu G, Chapman JR, Brandsma I, Yuan J, et al. REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature. 2015;521:541–4. doi: 10.1038/nature14328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aly A, Ganesan S. BRCA1, PARP, and 53BP1: conditional synthetic lethality and synthetic viability. J Mol Cell Biol. 2011;3:66–74. doi: 10.1093/jmcb/mjq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jaspers JE, Kersbergen A, Boon U, Sol W, et al. Loss of 53BP1 causes PARP inhibitor resistance in Brca1-mutated mouse mammary tumors. Cancer Discov. 2013;3:68–81. doi: 10.1158/2159-8290.CD-12-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bashyam MD. Nonsense-mediated decay: linking a basic cellular process to human disease. Expert Rev Mol Diagn. 2009;9:299–303. doi: 10.1586/erm.09.18. [DOI] [PubMed] [Google Scholar]
- 71.Miller JN, Pearce DA. Nonsense-mediated decay in genetic disease: friend or foe? Mutat Res Rev Mutat Res. 2014;762:52–64. doi: 10.1016/j.mrrev.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE. Nonsense-mediated decay approaches the clinic. Nat Genet. 2004;36:801–8. doi: 10.1038/ng1403. [DOI] [PubMed] [Google Scholar]
- 73.Finkel RS. Read-through strategies for suppression of nonsense mutations in Duchenne/ Becker muscular dystrophy: aminoglycosides and ataluren (PTC124) J Child Neurol. 2010;25:1158–64. doi: 10.1177/0883073810371129. [DOI] [PMC free article] [PubMed] [Google Scholar]