Fig. 1. Design of the feature probes of the rice alternatively spliced transcript detection microarray (ASDM).
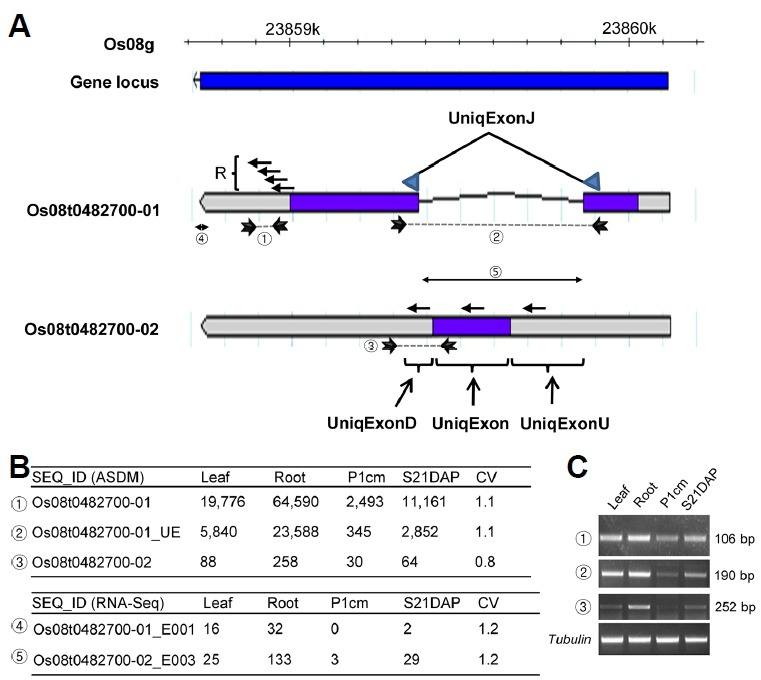
(A) Probes are positioned and over-scaled based on the genome features in RAP-DB (rapdb.dna.affrc.go.jp). Four probes are collectively indicated as R for the locus Os08g0482700 covering 60 bp of the CDS and 90 bp of the 3′ UTR. Additionally, a 60-bp feature probe (UniqExonJ) was designed by joining 30-bp-long spliced transcripts from two regions (arrow heads) in the CDS for each exon for Os08t0482700-01. Thus, the probe represents a unique junction of two exons of Os08t0482700-01 and is indicated as Os08t0482700-01_UE in the intensity file. The Os08t0482700-02 transcript has three unique regions compared to Os08t0482700-01: part of the 5′ UTR, the CDS and part of a 3′ UTR, designated as UniqExonU, UniqExon, and UniqExonD, respectively. Three 60-bp-long target probes were designed according to these regions. (B) Signal intensities and transcript numbers for rice ASDM (top) and RNA-Seq (bottom), respectively. Exons in RNA-Seq are designated according to the exon partition by dexseq_prepare_annotation.py (Anders et al., 2012). Exon bins are indicated by suffixing E and numeral generated by exonic_part_number attribute (Supplementary Table S4). (C) Confirmation of expression by semi-quantitative RT-PCR.
