Abstract
Background
DL-3-n-butylphthalide (NBP) is a drug for treating acute ischemic stroke, and may play a neuroprotective role by acting on multiple active targets. The aim of this study was to predict the target proteins of NBP in mammalian cells.
Methods
The similarity ensemble approach search tool (SEArch), one of the commonly used public bioinformatics tools for target prediction, was employed in the experiment. The molecular docking of NBP to target proteins was performed by using the three-dimensional (3-D) crystal structure, substrate free. The software AutoDock Vina was used for all dockings. The binding targets of NBP were illustrated as 3-D and 2-D diagrams.
Results
Firstly, the results showed that NBP bounded to the same binding site on NAD(P)H quinone oxidoreductases (NQO1) as the substrate FAD, leading to competitive inhibition for the catalytic site with −7.2 kcal/mol. This might break the 3-D structure of NQO1 and bring about P53 degradation, resulting in a decrease of p53-mediated apoptosis in ischemic brain cells. Secondly, NBP might exert its therapeutic effect on acute ischemic stroke via modulating indoleamine 2,3-dioxygenase (IDO) bioactivity after associating with it. NBP could alleviate the depression following ischemic stroke by inhibiting IDO. Thirdly, NBP might modulate the function of NADH-ubiquinone oxidoreductase by competitively embedding itself into this complex, further affecting mitochondrial respiration in cerebrovascular diseases as an anti-oxidant agent.
Conclusions
Three potential target proteins of NBP were identified, which may provide a novel aspect for better understanding the protective effects of NBP on the nervous system at the molecular level.
Keywords: DL-3-n-butylphthalide (NBP); molecular targets; bioinformatics; NAD(P)H quinone oxidoreductases; indoleamine 2,3-dioxygenase (IDO); NADH-ubiquinone oxidoreductase
Introduction
DL-3-n-butylphthalide (NBP) is the first class I novel drug of China used in the cerebrovascular field with an independent intellectual property right. Since it was approved in 2002, NBP has only been used for treating acute ischemic stroke. Recently, studies also show that NBP has a therapeutic effect on various neurodegenerative diseases, especially vascular dementia and vascular cognitive impairment without dementia (1). In animal experiments, including our own investigations, NBP may reduce cerebral edema, decrease cerebral infarct volume, improve cerebral microcirculation, protect vascular endothelial cells, scavenge free radicals, protect mitochondria, reduce inflammation, inhibit neuronal apoptosis, counteract platelet aggregation, inhibit thrombosis and decrease the expression of β-amyloid protein and tau protein in the brain (1-3). Therefore, NBP may play a neuroprotective role by acting on multiple active targets.
However, there is a dearth of studies on the molecular targets of NBP. Identification of its molecular targets is not only the key to further reveal the molecular mechanism of its neuroprotective effect, but also a source providing theoretical clues for developing new therapeutic indications. In this experiment, we explored the potential targets of NBP by employing the method of similarity ensemble approach (SEA) in combination with molecular docking. This study may open the door to investigate the molecular mechanism of the neuroprotective effect of NBP.
Methods
Target prediction by similarity ensemble approach search tool (SEArch)
Primarily, NBP’s target proteins in mammalian cells were predicted by using a SEArch, one of the commonly used public bioinformatics tools for target prediction based on the similarity of binding ligands (http://sea.bkslab.org/search/). The SMILE string of NBP, O=C1OC(c2ccccc12)CCCC was inputted into SEArch. The potential targets of NBP were ranked by E-values and MaxTC.
Molecular docking
Based on SEA prediction, molecular docking of NBP to target proteins was performed using the three-dimensional (3-D) crystal structure, substrate free. NAD(P)H quinone oxidoreductases (PDB code 1D4A), indoleamine 2,3-dioxygenase (IDO) (PDB code 2D0T) and NADH-ubiquinone oxidoreductase (PDB code 2YBB) were firstly obtained from the Protein Data Bank (http://www.rcsb.org/pdb/home/home.do). The following molecular docking studied was carried out by software AutoDock Vina (v.1.0.2). The docking parameters for AutoDock Vina were kept at their default values. The binding modes were clustered through the root-mean square deviation (RMSD) among the Cartesian coordinates of the ligand atoms. The docking results were ranked according to their binding free energies. The binding modes with the lowest binding free energies and the most clustered members were chosen for optimum docking conformation. The binding results were illustrated as 3-D and two-dimensional (2-D) diagrams by PyMOL Molecular Graphics System Version 1.3 (Schrödinger, LLC) and Ligplot plus (EMBL-EBI), respectively.
Results
A series of NBP potential targets resulting from SEArch are shown in Table 1 with different E-values and MaxTC. According to the SEA algorithm, the smaller the E-value, the closer the NBP-target interaction (4). All the targets, NAD(P)H quinone oxidoreductases, IDO and NADH-ubiquinone oxidoreductase were subjected to the following molecular docking study.
Table 1. Predicated protein targets of NBP by SEA analysis.
| Code | Ligands | Reference name | E-value | MaxTe |
|---|---|---|---|---|
| NQO1_HUMAN_10000 | 79 | NAD(P)H quinone oxidoreductase | 6.43×10−9 | 0.32 |
| NU4LM_HUMAN_10000 | 11 | NADH-ubiquinone oxidoreductase chain 4L | 2.54×10−3 | 0.30 |
| I23O1_HUMAN_10000 | 69 | Indoleamine 2,3-dioxygenase | 1.63×10−2 | 0.31 |
| NU2M_HUMAN_10000 | 18 | NADH-ubiquinone oxidoreductase chain 2 | 1.04×10−1 | 0.30 |
| NU3M_HUMAN_10000 | 18 | NADH-ubiquinone oxidoreductase chain 3 | 1.04×10−1 | 0.30 |
| NU6M_HUMAN_10000 | 18 | NADH-ubiquinone oxidoreductase chain 6 | 1.04×10−1 | 0.30 |
| NU1M_HUMAN_10000 | 18 | NADH-ubiquinone oxidoreductase chain 1 | 1.04×10−1 | 0.30 |
| NU5M_HUMAN_10000 | 18 | NADH-ubiquinone oxidoreductase chain 5 | 1.04×10−1 | 0.30 |
| NU4M_HUMAN_10000 | 23 | NADH-ubiquinone oxidoreductase chain 4 | 4.42×10−1 | 0.30 |
SEA, similarity ensemble approach; NBP, DL-3-n-butylphthalide.
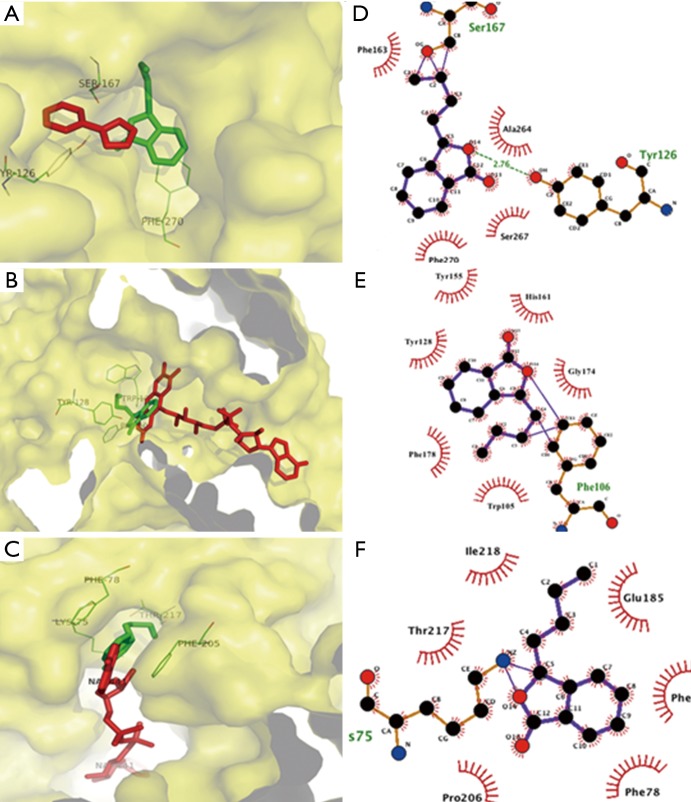
The results of molecular docking study showed that NBP bounded to the same binding site on NAD(P)H quinone oxidoreductases as the substrate FAD, ensuing in competitive inhibition for the catalytic site with −7.2 kcal/mol. NBP interacted with Trp105, Phe178, Tyr128, Tyr155, His161 and Gly174 with hydrophobic bonds, as well as Phe106 possessing external covalent bonds, thus interrupted FAD binding to specific residues and triggering of downstream signals. The 1,4-dihydronicotinamide adenine dinucleotide binding to NADH-ubiquinone oxidoreductases served as a key role in mitochondrial supercomplex function, which can be inhibited by NBP through binding to the same pocket with −7.7 kcal/mol. Docking results illustrated that NBP can form an external covalent bond with Lys75 and hydrophobic bonds with Thr217, Ile218, Glu185, Phe120, Phe78 and Pro206. Besides binding to mitochondrial complex, NBP also targeted at the inhibitor-binding site of IDO with −7.5 kcal/mol. Ser167 and Tyr126 interacted with NBP through an external covalent bond and a hydrogen bond, respectively (Figure 1).
Figure 1.
Molecular docking study showing the most energetically favorable binding three dimensional (3D) modes of NBP at the substrate-binding gorge of (A) NAD(P)H quinone oxidoreductase and (B) NADH-ubiquinone oxidoreductase and (C) inhibitor-binding indoleamine 2,3-dioxygenase. The red stick represents substrate or inhibitor, the green stick represents NBP, the yellow curved surface represents binding pocket surface and the green lines with numbered amino acids show the residues interacting with ligands. The two-dimensional (2-D) diagrams display the drug-protein interaction for (D) NAD(P)H quinone oxidoreductase, (E) NADH-ubiquinone oxidoreductase and (F) inhibitor-binding indoleamine 2,3-dioxygenase. The color of each residue represents the types of interactions as follows: hydrophobic bonds (brick red); external bonds (purple); hydrogen bonds (olive green). NBP, DL-3-n-butylphthalide.
Discussion
NAD(P)H quinone oxidoreductases (NQO1) is a 2-eletron reductase located within the cytosol, belonging to the NAD(P)H dehydrogenase (quinone) family. It is generally considered as an efficient detoxifying enzyme by transforming reactive factors such as quinones, quinoneimine and glutathionyl-substituted into harmless forms when cells are responding to a large spectrum of oxidative stresses (5). Moreover, NQO1 displays a controversial effect on cell survival and death: NQO1 can stabilize p53, which is a crucial tumor suppressor in certain human carcinomas; NQO1 also enhances γ-irradiation-induced apoptosis in normal thymocytes, which is p53-dependent (6). On the other hand, animal experiment has indicated that NBP could reduce glial apoptosis in diabetic rat (7). As in our study, one potential mechanism of the clinical effects of NBP on neurological functions is that competitive binding of NBP to NQO1 may break the 3-D structure of NQO1 and cause P53 degradation, resulting in a decrease of p53-medicated apoptosis in ischemic brain cells. Hence, it is possible that NBP suppresses glial apoptosis by restricted P53 degradation via inhibition of NAD(P)H quinone oxidoreductases.
IDO is a key enzyme in kynurenine pathway to catalyze the conversion of L-tryptophan to N-formylkynurenine. IDO is well-known for its checkpoint role in the immune system. Abnormal excessive IDO is frequently observed in a variety of diseases (8). Intriguingly, it has been demonstrated that IDO can modulate vascular tone and play an important role in cerebral ischemia-reperfusion in a mouse model (9). However, the underlying mechanisms are still unclear. Besides that, it is also reported that excessively expressed IDO could lead to depression following ischemic stroke (10,11). Moreover, over-expressed IDO in hippocampus may also play a role as a pathological factor in Alzheimer disease (12). In the paper our team published in 2012, it has already been shown that NBP also exerts the therapeutic effect in vascular dementia (3), which may be partially explained by previously reported functions of IDO. In conclusion, NBP may play a role as an anti-stroke and anti-dementia drug via inhibiting IDO.
As ubiquitous energy organelles, mitochondria provide most of the adenosine triphosphate (ATP) for cells through the respiratory chain comprising several complex and supercomplex (13). NADH-ubiquinone oxidoreductase is a complex containing 44 subunits. Disassociation or mutation of this complex exists within many human disorders such as Leigh syndrome (14). Of note, pathological defects of mitochondrial respiration have a close correlation with both acute neuronal trauma and chronic neurodegenerative diseases (13). Metformin, as an inhibitor of NADH-ubiquinone oxidoreductase, is proved to exert protective role in ischemic stroke through suppression of ROS (reactive oxygen species). NBP may suppress ROS by the mechanism relying on NADH-ubiquinone oxidoreductase in a manner of resembling metformin (15,16). Based on the paper we published in 2012 (2), NBP may prevent neurological deficits and cerebral injury following stroke as an anti-oxidant agent, which is in good agreement with our prediction. As NADH-ubiquinone oxidoreductase locates in the inner layer of mitochondria which is highly enriched in glial cells, it could hold true that the anti-ROS role of NBP relies on glial cells rather than neurons. Based on these studies and our data, we come to the hypothesis that NBP may modulate the function of NADH-ubiquinone oxidoreductases by competitively embedding itself into this complex, further affecting mitochondrial respiration in cerebrovascular diseases.
Our results are encouraging, but some limitations are worth noting. Although the results of molecular docking are in good agreement with SEA analysis, our data should be further validated. The future study utilizing molecular dynamics simulation and in-vitro binding assays could be performed to further confirm the predicted NBP binding sites.
In sum, our study firstly identifies three potential target proteins of NBP via SEA analysis in combination with molecular docking, which provides a novel aspect for a better understanding of the protective effects of NBP on the nervous system at the molecular level. These three target proteins themselves have extremely physical and pathological functions and may be able to serve as clinical targets of NBP in the future.
Acknowledgements
Funding: The authors gratefully thank the Key Project of Medical Science Research of Hebei Province, China (No. 20160569) and CSPC NBP Pharmaceutical Co., Ltd. for their generous support.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Abdoulaye IA, Guo YJ. A Review of Recent Advances in Neuroprotective Potential of 3-N-Butylphthalide and Its Derivatives. Biomed Res Int 2016;2016:5012341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang L, Yu WH, Wang YX, Wang C, Zhao F, Qi W, Chan WM, Huang Y, Wai MS, Dong J, Yew DT. DL-3-n-Butylphthalide, an anti-oxidant agent, prevents neurological deficits and cerebral injury following stroke per functional analysis, magnetic resonance imaging and histological assessment. Curr Neurovasc Res 2012;9:167-75. 10.2174/156720212801618956 [DOI] [PubMed] [Google Scholar]
- 3.Zhang L, Lü L, Chan WM, Huang Y, Wai MS, Yew DT. Effects of DL-3-n-butylphthalide on vascular dementia and angiogenesis. Neurochem Res 2012;37:911-9. 10.1007/s11064-011-0663-3 [DOI] [PubMed] [Google Scholar]
- 4.Keiser MJ, Roth BL, Armbruster BN, Ernsberger P, Irwin JJ, Shoichet BK. Relating protein pharmacology by ligand chemistry. Nat Biotechnol 2007;25:197-206. 10.1038/nbt1284 [DOI] [PubMed] [Google Scholar]
- 5.Ross D, Siegel D. NAD(P)H:quinone oxidoreductase 1 (NQO1, DT-diaphorase), functions and pharmacogenetics. Methods Enzymol 2004;382:115-44. 10.1016/S0076-6879(04)82008-1 [DOI] [PubMed] [Google Scholar]
- 6.Asher G, Lotem J, Kama R, Sachs L, Shaul Y. NQO1 stabilizes p53 through a distinct pathway. Proc Natl Acad Sci U S A 2002;99:3099-104. 10.1073/pnas.052706799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang T, Jia W, Sun X. 3-n-Butylphthalide (NBP) reduces apoptosis and enhances vascular endothelial growth factor (VEGF) up-regulation in diabetic rats. Neurol Res 2010;32:390-6. 10.1179/016164110X12670144526264 [DOI] [PubMed] [Google Scholar]
- 8.Murakami Y, Hoshi M, Imamura Y, Arioka Y, Yamamoto Y, Saito K. Remarkable role of indoleamine 2,3-dioxygenase and tryptophan metabolites in infectious diseases: potential role in macrophage-mediated inflammatory diseases. Mediators Inflamm 2013;2013:391984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackman KA, Brait VH, Wang Y, Maghzal GJ, Ball HJ, McKenzie G, De Silva TM, Stocker R, Sobey CG. Vascular expression, activity and function of indoleamine 2,3-dioxygenase-1 following cerebral ischaemia-reperfusion in mice. Naunyn Schmiedebergs Arch Pharmacol 2011;383:471-81. 10.1007/s00210-011-0611-4 [DOI] [PubMed] [Google Scholar]
- 10.Mo X, Pi L, Yang J, Xiang Z, Tang A. Serum indoleamine 2,3-dioxygenase and kynurenine aminotransferase enzyme activity in patients with ischemic stroke. J Clin Neurosci 2014;21:482-6. 10.1016/j.jocn.2013.08.020 [DOI] [PubMed] [Google Scholar]
- 11.Ormstad H, Verkerk R, Amthor KF, Sandvik L. Activation of the kynurenine pathway in the acute phase of stroke and its role in fatigue and depression following stroke. J Mol Neurosci 2014;54:181-7. 10.1007/s12031-014-0272-0 [DOI] [PubMed] [Google Scholar]
- 12.Guillemin GJ, Brew BJ, Noonan CE, Takikawa O, Cullen KM. Indoleamine 2,3 dioxygenase and quinolinic acid immunoreactivity in Alzheimer's disease hippocampus. Neuropathol Appl Neurobiol 2005;31:395-404. 10.1111/j.1365-2990.2005.00655.x [DOI] [PubMed] [Google Scholar]
- 13.Vartak R, Porras CA, Bai Y. Respiratory supercomplexes: structure, function and assembly. Protein Cell 2013;4:582-90. 10.1007/s13238-013-3032-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ortigoza-Escobar JD, Oyarzabal A, Montero R, Artuch R, Jou C, Jiménez C, Gort L, Briones P, Muchart J, López-Gallardo E, Emperador S, Pesini ER, Montoya J, Pérez B, Rodríguez-Pombo P, Pérez-Dueñas B. Ndufs4 related Leigh syndrome: A case report and review of the literature. Mitochondrion 2016;28:73-8. 10.1016/j.mito.2016.04.001 [DOI] [PubMed] [Google Scholar]
- 15.Batandier C, Guigas B, Detaille D, El-Mir MY, Fontaine E, Rigoulet M, Leverve XM. The ROS production induced by a reverse-electron flux at respiratory-chain complex 1 is hampered by metformin. J Bioenerg Biomembr 2006;38:33-42. 10.1007/s10863-006-9003-8 [DOI] [PubMed] [Google Scholar]
- 16.Dröse S, Stepanova A, Galkin A. Ischemic A/D transition of mitochondrial complex I and its role in ROS generation. Biochim Biophys Acta 2016;1857:946-57. 10.1016/j.bbabio.2015.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]