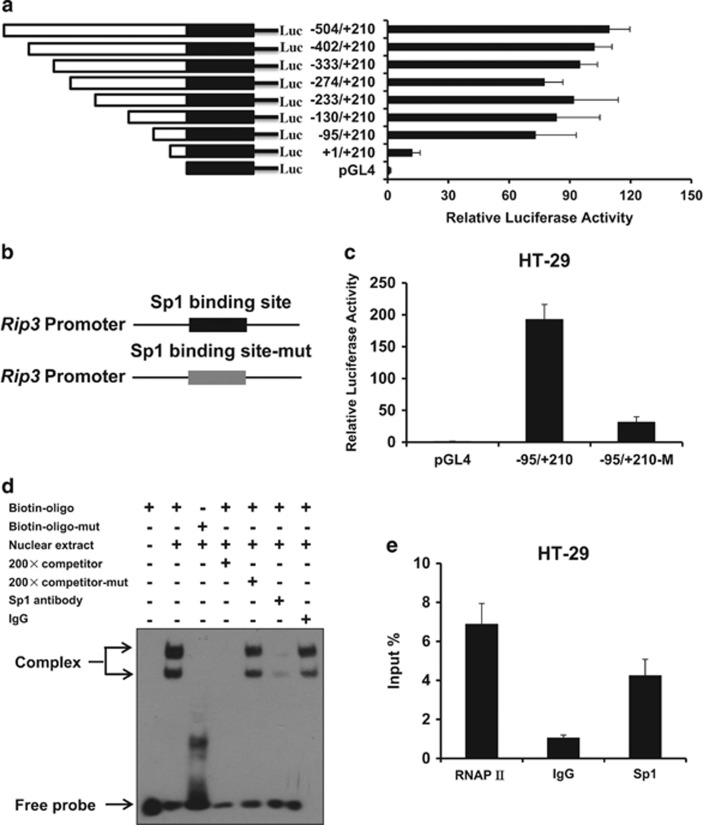
Figure 2.
Transcription factor Sp1 regulates RIP3 promoter activity. (a) Analysis for transcription activity of Rip3 promoter in HT-29 cells by the co-transfection of luciferase reporter plasmid (Rip3/pGL4 or pGL4) and the pRL-TK plasmid. The luciferase activity value of each sample was first normalized for transfection efficiency by co-transfection with the pRL-TK plasmid. The transcriptional activity of promoter construct was shown as the luciferase activity relative to that of the pGL4 vector (a promoter-less vector). (b) Schematic respectively represented WT and site-mutant Rip3 promoter. (c) Analysis for transcription activity of wild type or Sp1 binding site-mutant Rip3 promoter in HT-29 cells by the co-transfection of luciferase reporter plasmid and the pRL-TK plasmid. (d) The binding of transcription factor Sp1 to the Rip3 promoter was determined by electrophoretic mobility shift analysis (EMSA). By using biotin-labeled 30 bp double-stranded oligonucleotides containing wild type or mutated Sp1 binding sites as probes, EMSAs were performed with the same amount of nuclear extracts (10 μg) from HT-29 cells, and the products were separated on a 6% polyacrylamide gel (lanes 2–7). Lane 1, free probe; lane 2, biotin-labeled wild-type Sp1 consensus oligonucleotides were mixed with nuclear proteins; lanes 3, binding assays of biotin-labeled mutant-type Sp1 consensus oligonucleotides mixed with nuclear proteins; lane 4, the same reaction was performed as that in lane 2, except for the presence of a 200-fold excess of unlabeled wild-type Sp1 consensus oligonucleotides as a competitor; lane 5, the same reaction was performed as that in lane 2, except for the presence of a 200-fold excess of unlabeled mutant-type Sp1 consensus oligonucleotides as a competitor; lanes 6–7, IgG or anti-Sp1 antibody was added to the binding reaction mixtures with biotin-labeled wild-type probe. (e) ChIP assay using antibody against Sp1 was performed in HT-29 cells. The normal rabbit IgG was used as a negative control, the anti-polymerase-II antibody was used as a positive control and input indicates 5% input DNA, a positive amplification control