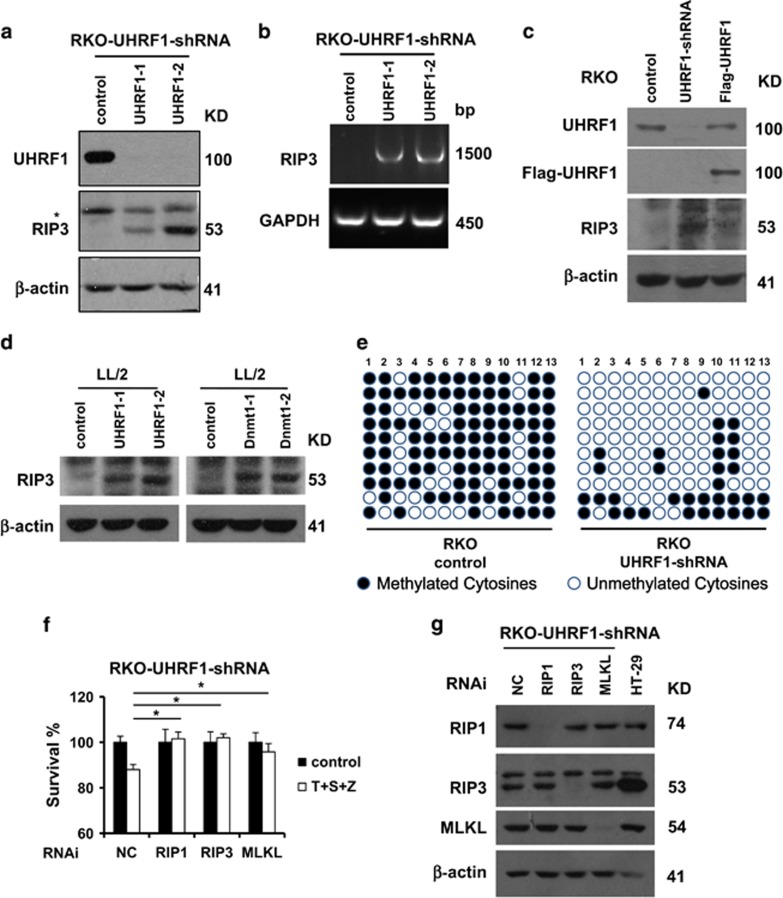
Figure 4.
The epigenetic regulator UHRF1 controls epigenetic silencing of RIP3 in cancer cells. (a) RKO cells, UHRF1-shRNA stable cell lines were generated as described in the Materials and Methods, and western blotting analysis of lysates from the indicated stable cell lines showing UHRF1, RIP3 and β-actin levels. RKO-UHRF1-shRNA: RKO cells stably expressing a shRNA targeting UHRF1. (b) Reverse transcription-PCR products from the indicate cells to detect the Rip3 mRNA. (c) Western blotting analysis of lysates from the indicated stable cell lines showing UHRF1, Flag, RIP3 and β-actin levels. Flag-UHRF1: UHRF1-shRNA cells transiently expressing an shRNA-resistant UHRF1. (d) RNAi of UHRF1 and Dnmt1 in LL/2 cells. Western blotting analysis of lysates from the indicated stable cell lines showing RIP3 and β-actin levels. (e) Methylation status of the CpG-dinucleotides of DNA sequences (−152 to +240 bp) upstream and downstream of RIP3 transcription start site was validated by bisulfite sequencing from the indicated cell lines. (f) RKO-UHRF1-shRNA cells were transfected with the control, RIP1 siRNA, RIP3 siRNA or MLKL siRNA. Forty-eight hours post transfection, cells were treated as indicated for additional 48 h. Cell viability was determined by measuring ATP levels. *P<0.05. (g) The knockdown efficiency of RIP1, RIP3 and MLKL RNAi. Cell lysates were collected 48 h post transfection and aliquots of 40 μg were subjected to western blot analysis of RIP1, RIP3, MLKL and β-actin levels. All experiments were repeated at least three times with similar results