Abstract
Chikungunya fever is a major public health issue in India affecting billions. After 2010, the infection was in a decline until in 2016, when a massive outbreak affected the country. In this report, we present serologic and molecular investigations of 600 patient samples for chikungunya and dengue viruses along with clinical and comorbidity features. We recruited 600 patients during this outbreak and evaluated them for chikungunya and dengue virus antibodies and virus RNA through IgM, NS1 antigen and quantitative real-time PCR (qPCR). We further evaluated Zika virus RNA by qPCR. Additionally, we documented all clinical and comorbid features that were observed during the outbreak in the hospital. We report a total incidence rate of 58% of chikungunya during the outbreak in our hospital. Within the recruited patients, 70% of the patients were positive for chikungunya virus IgM whereas 24.17% were positive by qPCR. None of the samples was positive for Zika virus RNA. Additionally, coinfection of dengue and chikungunya was seen in 25.33% of patients. Analysis of clinical features revealed that 97% of patients had restricted movements of the joints with other features like swelling, itching and rashes of varying severity observed. Twelve patients presented with comorbid conditions, and two fatalities occurred among these comorbid patients. The high incidence of coinfection in the current outbreak warrants implementation of routine testing of both chikungunya and dengue virus in suspected patients for better patient management. The post–acute phase complications reported in the hospitals require in-depth studies to understand the actual impact of the current outbreak.
Keywords: Chikungunya, India, outbreak, 2016
Introduction
India has been affected by chikungunya fever (CHIKF) every two to three decades since the 1960s [1]. The latest pandemic that occurred in several parts of the globe in 2005 resulted in huge outbreaks in India, resulting in huge loss of disability-adjusted life-years and a major public health concern in the country [2], [3], [4]. In 2010, Delhi witnessed an outbreak [5], and since then, CHIKF has been in a decline; only sporadic cases been reported. However, in August 2016, another massive outbreak broke in New Delhi and the National Capital Region, resulting in thousands of households being bedridden (http://nvbdcp.gov.in/chik-cd.html). The present report describes the status of the CHIKF outbreak in New Delhi during 2016, with an emphasis on the status of coinfections and comorbid conditions of recruited patients.
Methods
Between August and December 2016, a total of 5180 patients sought care at the Department of Microbiology and the outpatient wards of Safdarjung Hospital for suspected CHIKF. Of these, 600 patients were enrolled onto this study, which was part of an ongoing project that was cleared by the institutional ethical board (IEC/VMMC/SJH/Project/February-2016/575, ICGEB/IEC/2016/01 and Version-3). Blood samples were collected, and clinical information such as age, gender, symptoms, joint movement and onset of fever were documented. Sera were separated and stored at −80°C until further use. Admission criteria also included patients with complications and associated comorbid conditions. Cerebrospinal fluid was collected from two such patients and stored at −80°C. All sera samples were subjected to IgM serology for the presence of dengue virus (DENV) and chikungunya virus (CHIKV) using government-recommended kits from National Institute of Virology, Pune, NS1. Antigen for DENV was detected using Qualpro diagnostics. Quantitative real-time PCR (qPCR) for the presence of CHIKV RNA was performed using previously published primers [6], and Zika virus RNA was tested using qPCR kits from Altona Diagnostics. Cerebrospinal fluid was tested directly and was also inoculated in C6/36 cells to evaluate the presence of CHIKV through qPCR. qPCR-positive bands were eluted from gel, sequenced and phylogenetic analysis performed for the sequences.
Results and discussion
A total of 2982 patients tested positive for CHIKF out of the total 5180, resulting in an incidence rate of 58% during the collection period. Of these, demographic and laboratory data were collected for a total of 600 patients enrolled onto the study (Table 1). The median age of patients was 35 years (range, 11–68 years). Male-to-female ratio was 1.1:1.0. At the time of patient recruitment, onset of fever ranged from 1 to 30 days (median, 4 days).
Table 1.
Demographic and laboratory data
| Characteristic | Value |
|---|---|
| Demographic (n = 600) | |
| Age (years) | |
| Mean | 35 |
| Range | 11–68 |
| Sex, M:F | 1.1:1 |
| Onset of fever (days) | |
| Median | 4 |
| Range | 1–30 |
| Clinical (n = 150) | |
| Patients with: | |
| Fever with chills | 9 (6%) |
| Itching | 35 (23.3%), severe 2 (1.3%) |
| Nausea | 12 (8%) |
| Rashes | 46 (30.6%), severe 11 (7.3%), moderate 21 (14.0%) |
| Diarrhoea | 2 (1.3%) |
| Vomiting | 5 (3.3%) |
| Ulcers | 2 (1.3%) |
| Headache | 7 (5%) |
| Musculoskeletal | |
| Swelling | 59 (39%), severe 2 (1.3%) |
| Restricted movement of the joints | 146 (97%), extreme 41 (27%) |
| Backache | 3 (2%) |
| Morning stiffness | 32 (21.3) |
| Body ache | 3 (2%) |
| Pedal oedema | 22 (14.6%) |
| Partial paraparesis | 4 (2.6%) |
| Comorbid Conditions (n = 12) | |
| Hypotension with additional complications | 3 (2%) |
| Hypertonia | 2 (1.3%) |
| Decreased sensation of pain, touch and temperature | 2 (1.3%) |
| Lipoma in head | 2 (1.3%) |
| Chest pain | 2 (1.3%) |
| Encephalopathy and encephalitis | 2 (1.3%) |
| Dermatomyositis | 1 (0.5%) |
| Respiratory presentation | 1 (0.5%) |
| Severe thrombocytopenia | 1 (0.5%) |
| Demyelinating disorder presenting as traverse myelitis | 1 (0.5%) |
| Intrauterine death | 1 (0.5%) |
| Mortality rate | 2 (1.3%) |
| Diagnostics (n = 600) | |
| CHIKV IgM positive | 420 (70%) |
| CHIKV qPCR positive | 145 (24.17%) |
| CHIKV IgM and qPCR positive | 40 (6.67%) |
| Only CHIKV IgM positive (CHIKV qPCR negative) | 244 (40.67%) |
| Only CHIKV qPCR positive (CHIKV IgM negative) | 103 (17.17%) |
| DENV IgM positive | 168 (28%) |
| DENV NS1 positive | 3 (0.5%) |
| ZIKV qPCR | 0 |
| Coinfection Status (n = 600) | |
| CHIKV/DENV | |
| CHIKV qPCR/DENV IgM | 40 (6.67%) |
| CHIKV qPCR/DENV NS1 | 0 (0%) |
| CHIKV IgM/DENV IgM | 112 (18.67%) |
| CHIKV IgM/DENV NS1 | 1 (0.5%) |
| Overall coinfection (CHIKV IgM/qPCR/DENV IgM/NS1 | 152 (25.33%) |
CHIKV, chikungunya virus; DENV, dengue virus; qPCR, quantitative real-time PCR; ZIKV, Zika virus.
Serologic investigations of the samples revealed that 420 samples (70%) were positive for CHIKV IgM, 145 samples (24.17%) were positive by qPCR and 40 samples (6.67%) were positive by both methods. With respective to dengue, DENV IgM tested positive in 168 samples (28%), and only three samples tested positive for the NS1 antigen. On the basis of these investigations, it was observed that 152 samples (25.33%) tested positive for both CHIKV and DENV, as measured by qPCR, IgM or both. None of the samples tested positive for Zika virus RNA by quantitative qPCR.
Analysis of the clinical features in 150 patients revealed that 97% (n = 146) of patients had restricted movements of the joints, with 41 patients displaying extreme restriction and requiring help in even lifting their limbs (Table 1). Swelling in joints was observed in 39% of the patients, with severe swelling seen in two patients. Itching and rashes occurred in 23.3% (n = 35) and 30.6% (n = 46) of patients respectively, with variable levels of severity (Table 1). Details of clinical features, musculoskeletal involvement and comorbid conditions are documented in Table 1.
Complications in CHIKF during this outbreak were seen in 12 patients during the time of sample collection (Table 1). Of these, hypotension was seen in three patients, with additional complication such as abdominal tenderness, coinfection with dengue and hematemesis. Encephalopathy/encephalitis was seen in two patients. Other comorbid conditions are listed in Table 1. Cerebrospinal fluid that was collected from two patients with comorbid conditions was cultured and revealed positivity for CHIKV through qPCR.
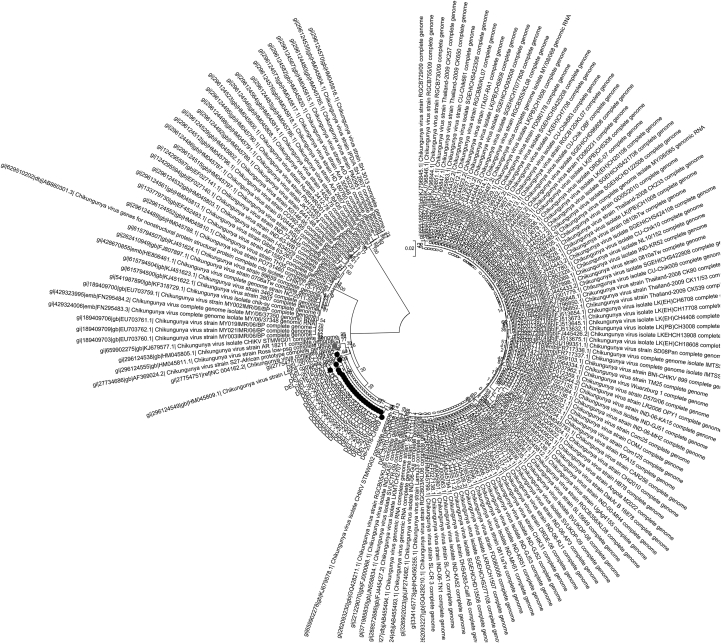
PCR products corresponding to complete gene of the structural envelope protein E1, from 21 samples including the cerebrospinal fluid samples, were purified and sequenced. Sequence analysis revealed that all samples belonged to Asian and East-Central South African (ECSA) genotype at the CHIKV E1 region (Fig. 1). Nucleotide sequence analysis performed on the whole E1 gene revealed variations at 40 nucleotide positions when compared to the 1953 Tanzanian strain (accession no. 045811.1). Of these, 29 variations resulted in synonymous and 11 variations in nonsynonymous mutations. At the amino acid level, 11 amino acid substitutions—namely, R123K, T145S, V179M, K211E, P232Q, I261D, M269V, D284E, I317V/M, V322A and N389K—were observed. Of these mutations, K211E, M269V and D284E have been reported previously in Delhi samples [5]. With respect to K211E, the mutation has been seen in all CHIKV samples after 2010 episode since its initial report in 2006 in a single sample in South India [7]. This mutation has been observed until 2014 in all reported samples [8]. However, for the first time, in the present study, two samples from the 2016 outbreak revealed reversion of wild-type lysine from the mutant glutamic acid. Considering that this amino acid site showed positive selection for the mutation in our previous study [5], in-depth analyses are warranted to understand the implication of this reversion in evolution of CHIKV and its impact in disease prevalence.
Fig. 1.
Genome sequence analysis of chikungunya virus (CHIKV) E1 region of all samples confirms that all samples belonged to Asian and East-Central South African (ECSA) genotype.
Conclusions
Since 2010, CHIKF had been declining in the country [9] until the current outbreak in 2016. However, chikungunya has been reported as a coinfection in almost 10% of DENV cases in recent studies [10]. The present outbreak reveals that the percentage is much higher, with 25% of CHIKF cases to be positive for DENV RNA antibodies, thereby making it imperative to check all suspected DENV cases for possible chikungunya infection as well. The current outbreak reports neurologic complications in CHIKF patients, as evidenced in a recent study on samples collected during the same outbreak in another hospital [11].
The chikungunya outbreak in 2016 has left long-lasting effects on the affected population. Post–acute phase complications are still being reported in the hospitals, and detailed analyses of the aftermath of the outbreak will be important to understand the actual impact of the current outbreak.
Acknowledgements
We thank all the patients enrolled onto this study for their agreement to participate. Supported in part by funding from research grants BT/PR14725/AGR/36/672/2010 (to SS) and DST/INT/JST/P-25/2014 (to RG and SS). Work in the AM laboratory was supported by grants PAR FSC 2007-2013 Flavipoc and LR 17/2014 Sevare from the Region FVG. MKV benefits from an Arturo Falaschi postdoctoral fellowship from the International Centre for Genetic Engineering and Biotechnology.
Conflict of Interest
None declared.
References
- 1.Shah K.V., Gibbs C.J., Jr., Banerjee G. Virological investigation of the epidemic of haemorrhagic fever in Calcutta: isolation of three strains of chikungunya virus. Ind J Med Res. 1964;52:676–683. [PubMed] [Google Scholar]
- 2.Mavalankar D., Shastri P., Raman P. Chikungunya epidemic in India: a major public-health disaster. Lancet Infect Dis. 2007;7:306–307. doi: 10.1016/S1473-3099(07)70091-9. [DOI] [PubMed] [Google Scholar]
- 3.Krishnamoorthy K., Harichandrakumar K.T., Kumari A.K., Das L.K. Burden of chikungunya in India: estimates of disability adjusted life years (DALY) lost in 2006 epidemic. J Vector Borne Dis. 2009;46:26–35. [PubMed] [Google Scholar]
- 4.Jain J., Nayak K., Tanwar N., Gaind R., Gupta B., Shastri J.S. Clinical, serological and virological analysis of 572 chikungunya patients during the years 2010–2013 from India. Clin Infect Dis. 2017 doi: 10.1093/cid/cix283. [In press] [DOI] [PubMed] [Google Scholar]
- 5.Shrinet J., Jain S., Sharma A., Singh S.S., Mathur K., Rana V. Genetic characterization of Chikungunya virus from New Delhi reveal emergence of a new molecular signature in Indian isolates. Virol J. 2012;9:100. doi: 10.1186/1743-422X-9-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niyas K.P., Abraham R., Unnikrishnan R.N., Mathew T., Nair S., Manakkadan A. Molecular characterization of chikungunya virus isolates from clinical samples and adult Aedes albopictus mosquitoes emerged from larvae from Kerala, South India. Virol J. 2010;7:189. doi: 10.1186/1743-422X-7-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krishnamoorthy N., Kamarajs T., Joseph R., Jambulingam P. Genotyping of virus involved in the 2006 chikungunya outbreak in South India (Kerala and Puducherry) Curr Sci. 2007;93:1412–1416. [Google Scholar]
- 8.Singh P., Sharma P., Kumar S., Mala C., Rizvi M.A., Mittal V. Continued persistence of ECSA genotype with replacement of K211E in E1 gene of chikungunya virus in Delhi from 2010 to 2014. Asian Pacific J Trop Dis. 2016;6:564–566. [Google Scholar]
- 9.Muniaraj M. Fading chikungunya fever from India: beginning of the end of another episode? Ind J Med Res. 2014;139:468. [PMC free article] [PubMed] [Google Scholar]
- 10.Londhey V., Agrawal S., Vaidya N., Kini S., Shastri J.S., Sunil S. Dengue and chikungunya virus co-infections: the inside story. J Assoc Physicians India. 2016;64:36–40. [PubMed] [Google Scholar]
- 11.Agarwal A., Vibha D., Srivastava A.K., Shukla G., Prasad K. Guillain-Barre syndrome complicating chikungunya virus infection. J Neurovirol. 2017;23:504–507. doi: 10.1007/s13365-017-0516-1. [DOI] [PubMed] [Google Scholar]