Figure 1. Schematic of CRISPR/Cas9-mediated gene inactivation.
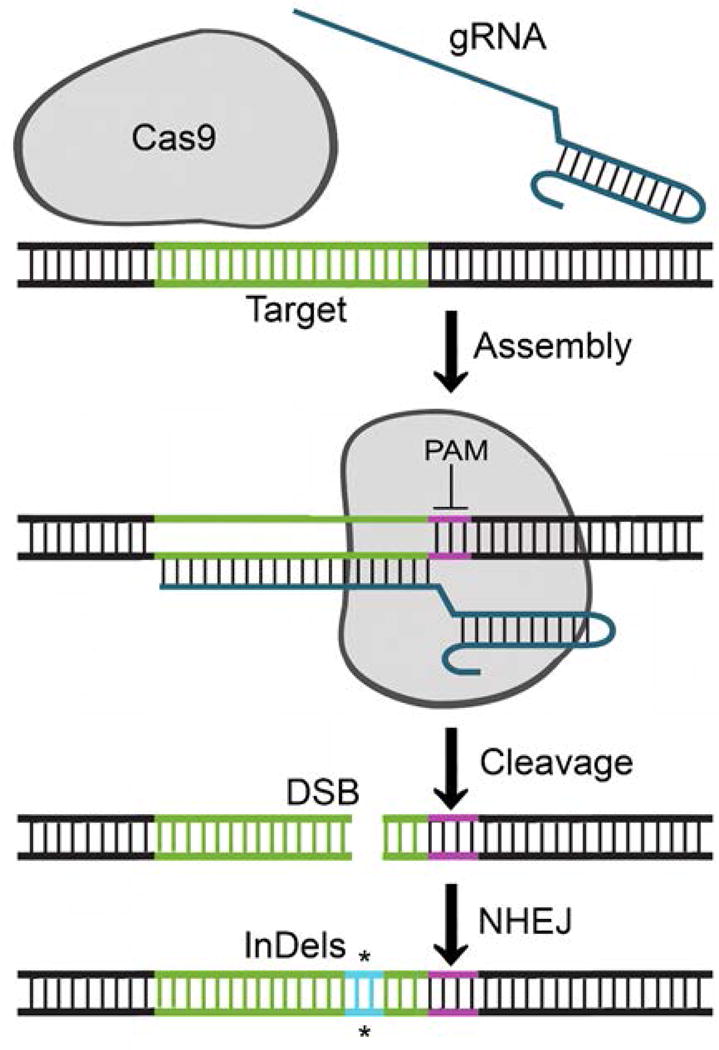
The schematic shows the action of Cas9 endonuclease on its target DNA (shown in green) and target-specific gRNA (shown in dark blue). In the first step, Cas9 binds to the target DNA guided by its complementarity to the DNA target and the presence of the PAM sequence. PAM is the protospacer adjacent motif and is shown in purple. In the second step, the target DNA undergoes endonucleolytic cleavage catalyzed by Cas9 to give a double-strand break (DSB). In the third step, the DSB is repaired by nonhomologous end-joining (NHEJ), which is an error-prone form of DNA repair that may allow the introduction of Insertion/Deletion mutations (InDels) that can impair the function of the gene (shown in bright blue and marked *).
