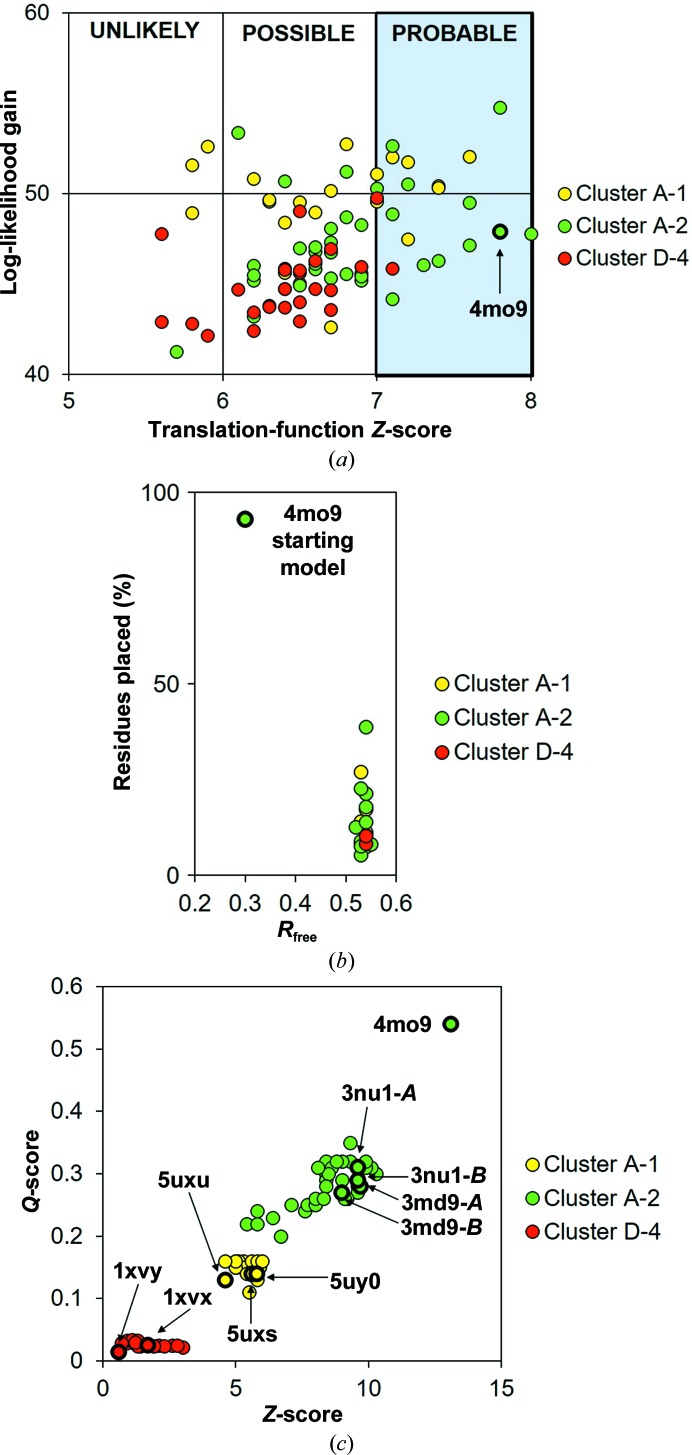
Figure 2.
Structure determination of YiuA and SBP alignment. (a) Scatterplot of MR scores from a brute-force search using cluster A-1, A-2 and D-4 SBP search models. Several search models achieved MR scores that suggest a probable solution, represented by data points in the shaded panel. PDB entry 4mo9, the only search model that successfully phased the YiuA data, is notated. (b) Brute-force automated model building using all starting models that achieved probable MR scores. Only the starting model from the 4mo9 search built >50% of the YiuA amino acids. (c) Secondary-structural alignment of YiuA against all SBPs used in (a). Alignments are scored by PDBeFold Q-score and Z-score algorithms. PDB entry 4mo9 is the most structurally similar SBP to YiuA of the test set. Other Y. pestis SBPs are labeled as well: YfeA (PDB entries 5uxs, 5uxu and 5uy0), HmuT (PDB entries 3md9 and 3nu1) and YfuA (PDB entries 1xvx and 1xvy). The HmuT results are also denoted with A for molecule A and B for molecule B.