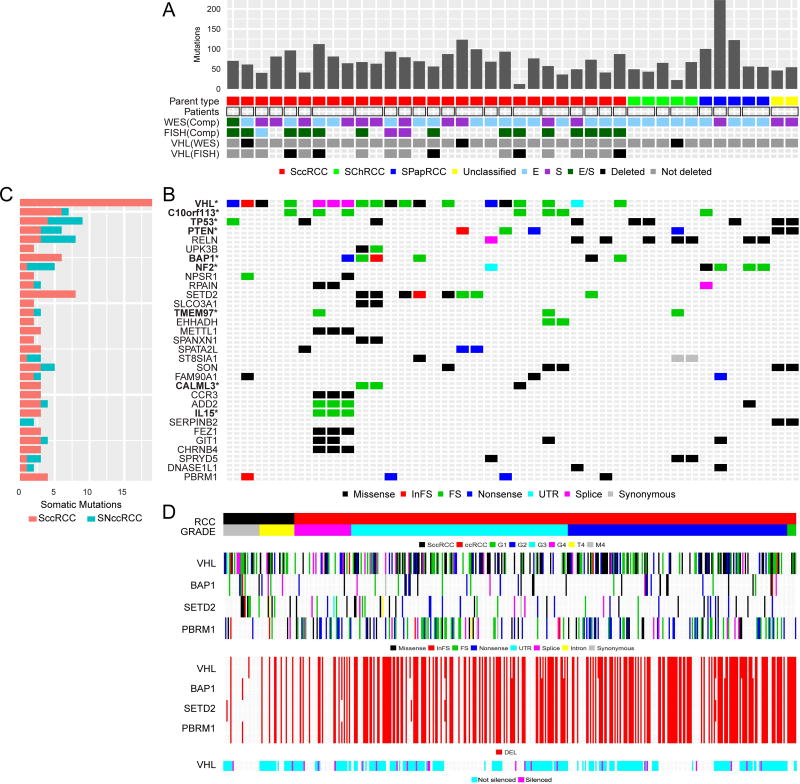
Figure 1. Mutational, copy number and methylation landscape of sarcomatoid RCC.
(A) Mutation frequency sorted by parent RCC subtype (SccRCC, SPapRCC, SChRCC, and Unclassified); unique patient samples indicated by square borders with paired patient samples indicated by rectangular borders; E- and/or S- components from each patient assayed by whole exome sequencing (WES) or Fluorescence in situ hybridization (FISH) with VHL deletions assessed by WES and FISH. (B) Types of somatic mutations involving specific genes, including significantly mutated genes by mutation frequency and found by MutSig*. (C) Bar plot shows mutations grouped by sarcomatoid clear cell (SccRCC) or sarcomatoid non-clear cell RCC (SNccRCC) subtype. (D) Sarcomatoid clear cell RCC derived from MD Anderson (M4) and TCGA (T4) cohorts and clear cell RCC (Grades 1–4) from TCGA in terms of the “hits” on 3p genes (VHL, PBRM1, SETD2, BAP1) consisting of mutations, deletions (DEL) and epigenetic silencing by methylation.