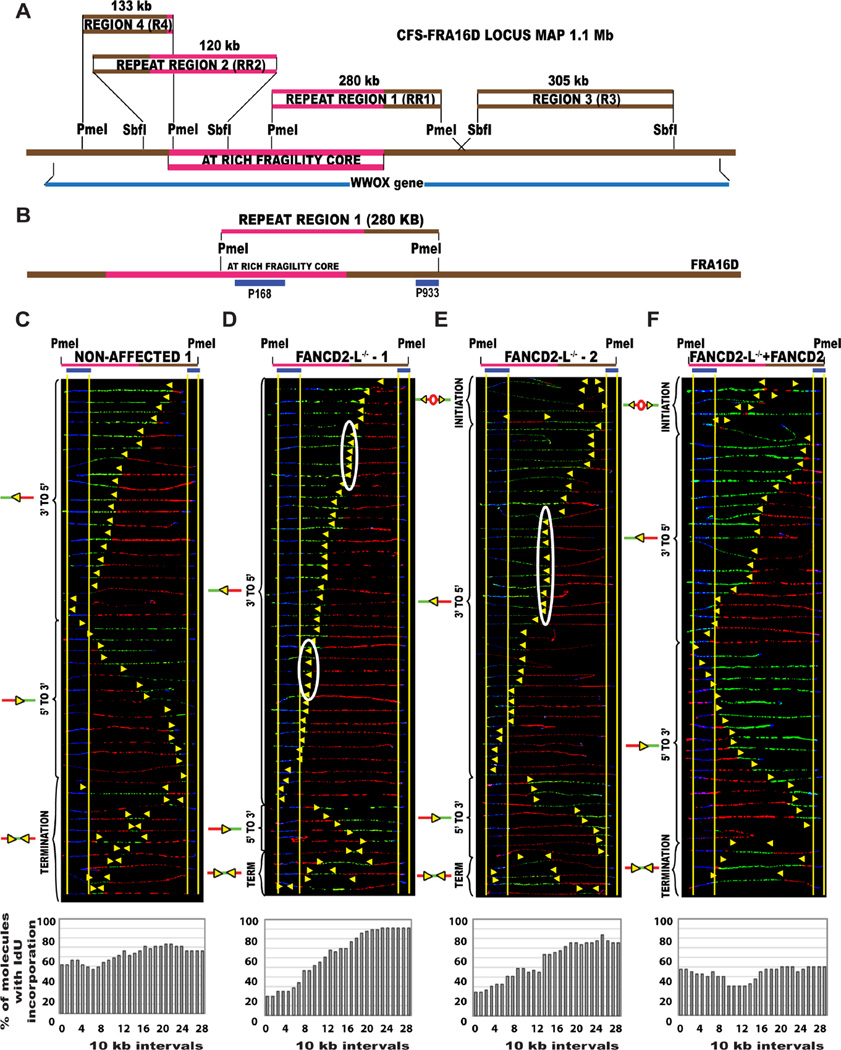
Figure 1. The replication profile is altered at the endogenous CFS-FRA16D locus in the absence of FANCD2.
(A) Common fragile site FRA16D locus map Locus map of the CFS-FRA16D (brown line-1.5 Mb) that contains the AT-rich fragility core (pink line–280 kb) and overlaps the WWOX tumor suppressor gene (dark blue line–1.1 Mb). The locus was divided into 4 segments based on restriction enzyme availability. The coordinates of the different regions are summarized in Table S1, providing additional information about fosmids and primers used to identify the regions.
(B) Locus map of RR1-PmeI segment containing a portion of the AT-rich fragility core. The segments are aligned according to the positions of the FISH probes (blue) on the map.
(C–F) Top; Locus map of PmeI digested RR1 segment. Middle; Aligned photomicrograph images of labeled DNA molecules from (C) Non-affected 1 (GM02184), (D) FANCD2−/−-L-1 (PD20), (E) FANCD2−/−-L-2 (2742) and (F) FANCD2−/−-L-1 + FANCD2 (corrected) lymphoblast. The yellow arrows indicate the sites along the molecules where the IdU transitioned to CldU. The molecules are arranged in the following order: molecules with initiation events, molecules with 3’ to 5’ progressing forks, molecules with 5’ to 3’ progressing forks and molecules with termination events. White ovals indicate regions of replication fork pausing and correspond to the pausing peaks listed in Table S2. Bottom; The percentage of molecules incorporating IdU (red) is calculated from the replication program (middle) and is represented as a histogram.