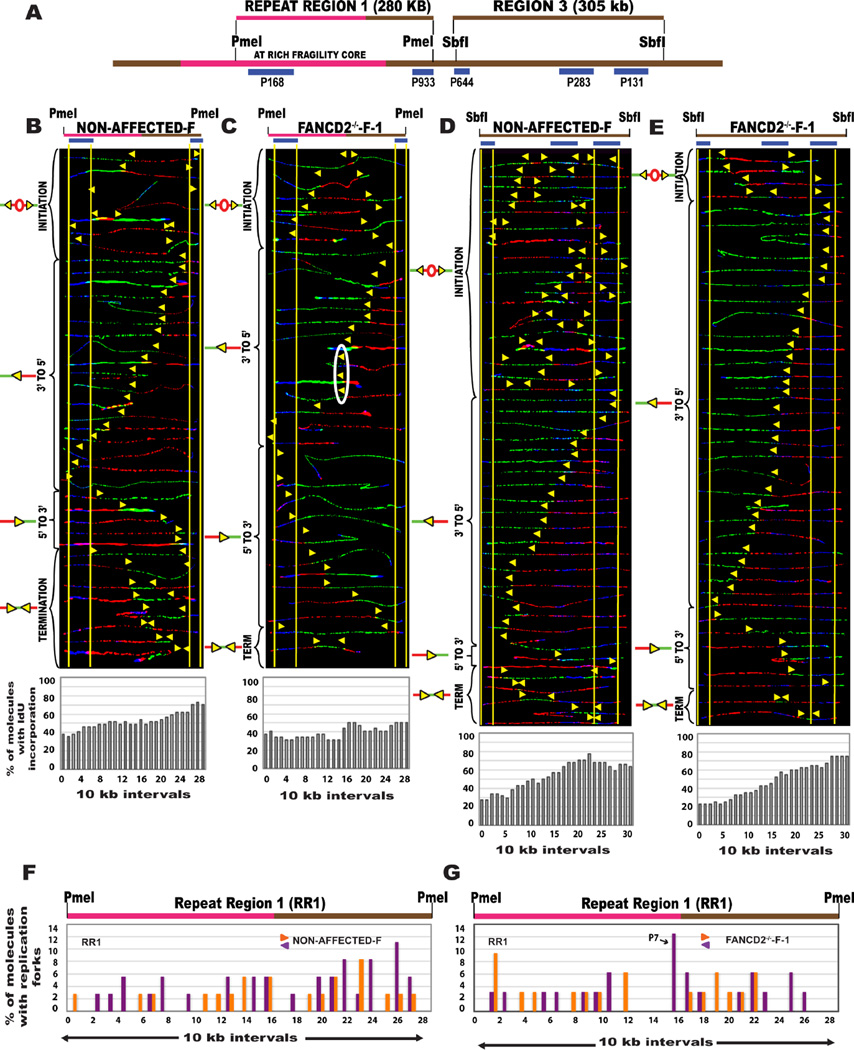
Figure 4. The absence of FANCD2 protein affects the replication program of CFS-FRA16D in lymphoblasts but not in fibroblasts.
(A) Locus map of the RR1-PmeI and R3-SbfI segments. The FISH probes that identify the segment are labeled in blue.
(B–C) Top; Locus map of PmeI digested RR1 segment. Middle; Aligned photomicrograph images of labeled DNA molecules from (B) Non-affected-F (IMR90 fibroblast), (C) FANCD2−/−-F-1 (PD20) patient fibroblasts. The yellow arrows indicate the sites along the molecules where the IdU transitioned to CldU. The molecules are arranged as in Fig. 2. Bottom; The percentage of molecules incorporating IdU (red) is represented as a histogram.
(D–E) Top; Locus map of SbfI digested R3 segment. Middle; Aligned photomicrograph images of labeled DNA molecules from (D) Non-affected-F (IMR90 fibroblast), (E) FANCD2−/−-F-1 (PD20) patient fibroblasts.
(F–G) Top; Locus map of the RR1 region. Bottom; The percentage of molecules with replication forks at each 10 kb interval of RR1 (quantification of molecules shown in Fig. 4B–C) in (F) Non-affected-F (IMR90 fibroblast), (G) FANCD2−/−-F-1 (PD20) patient fibroblasts. Black arrows denote the most prominent pause peaks and correspond to the white ovals in the SMARD profile.