Figure 6. The σ70 W-dyad and 6S RNA.
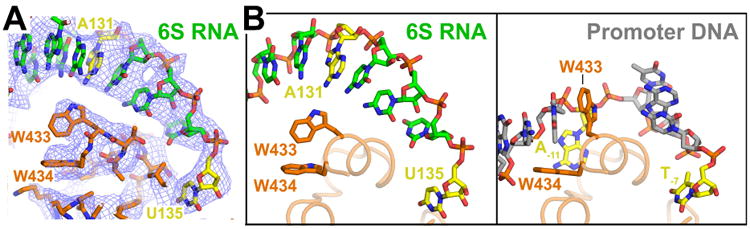
(A) View of cryoEM density (blue mesh) showing RNA nucleotide A131 (yellow) in a duplex RNA base stack passing over the σ70 W-dyad (absolutely conserved W433/W434). The Trp side chains maintain their edge-on conformation characteristic of σ70 alone (Malhotra et al., 1996) when not interacting with the double-strand/single-strand fork at the upstream edge of the transcription bubble (Bae et al., 2015).
(B) Comparison of 6S RNA (left) and promoter DNA (right) interacting with the absolutely conserved σ70 W-dyad (σ70 W433/W434). Shown is only the ‘nt-strand’ of the RNA or DNA. Corresponding nucleotides A131/U135 of 6S RNA or conserved A-11/T-7 of the promoter -10 element (Shultzaberger et al., 2007) are colored yellow. In both the RNA and DNA complexes, U135/T-7 is single-stranded and interacting with a pocket of σ70 (Feklistov and Darst, 2011). In the promoter DNA structure, the critical A-11 nucleotide is flipped out of the duplex base stack and σ70-W433 takes its place, forming a ‘chair’ conformation with W434 to stabilize the upstream edge of the newly formed transcription bubble (Bae et al., 2015). A131 remains in the base stack and the W-dyad maintains the edge-on conformation characteristic of σ70 alone (Feklistov and Darst, 2011). The 6S RNA is pre-melted (Figure 1A) and thus this interaction is dispensable.
See also Figure S6.
