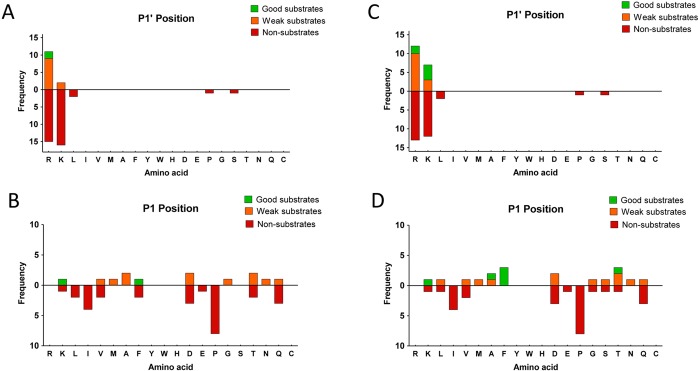
Fig 7. Analysis of the substrate preferences of CPD domains I and II using a tryptic peptide library.
(A,B) Substrate preferences of CPD domain I (i.e., rhCPD E762Q). (A) C-terminal P1’ position preferences. (B) P1 position preferences. (C,D) Substrate preferences of CPD domain II (i.e., rhCPD E350Q). (C) C-terminal P1’ position preferences. (D) P1 position preferences. The number of times each amino acid was present in P1 or P1’ was determined for good substrates, weak substrates and non-substrates. For P1 analysis, only substrates with permissive P1’ residues (Lys, Arg) were considered.