Abstract
Callose is a plant-specific polysaccharide (β-1,3-glucan) playing an important role in angiosperms in many developmental processes and responses to biotic and abiotic stresses. Callose is synthesised at the plasma membrane of plant cells by callose synthase (CalS) and, among others, represents the main polysaccharide in the callose wall surrounding the tetrads of developing microspores and in the growing pollen tube wall. CalS proteins involvement in spore development is a plesiomorphic feature of terrestrial plants, but very little is known about their evolutionary origin and relationships amongst the members of this protein family. We performed thorough comparative analyses of callose synthase family proteins from major plant lineages to determine their evolutionary history across the plant kingdom. A total of 1211 candidate CalS sequences were identified and compared amongst diverse taxonomic groups of plants, from bryophytes to angiosperms. Phylogenetic analyses identified six main clades of CalS proteins and suggested duplications during the evolution of specialised functions. Twelve family members had previously been identified in Arabidopsis thaliana. We focused on five CalS subfamilies directly linked to pollen function and found that proteins expressed in pollen evolved twice. CalS9/10 and CalS11/12 formed well-defined clades, whereas pollen-specific CalS5 was found within subfamilies that mostly did not express in mature pollen vegetative cell, although were found in sperm cells. Expression of five out of seven mature pollen-expressed CalS genes was affected by mutations in bzip transcription factors. Only three subfamilies, CalS5, CalS10, and CalS11, however, formed monophyletic, mostly conserved clades. The pairs CalS9/CalS10, CalS11/CalS12 and CalS3 may have diverged after angiosperms diversified from lycophytes and bryophytes. Our analysis of fully sequenced plant proteins identified new evolutionary lineages of callose synthase subfamilies and has established a basis for understanding their functional evolution in terrestrial plants.
Introduction
The colonisation of land by the ancestors of embryophytes represents the most significant event in the evolutionary history of terrestrial plants. Many key innovations or adaptations have appeared. The multi-layered pollen wall evolved to protect free-existing male gametophytes against environmental stress, which became critical when plants first colonised the terrestrial environment 600–450 million years ago. Algae, however, do not have complex spore walls [1]. The spores of ‘lower’ spore-bearing plants and the pollen of ‘higher’ seed plants are considered homologous [2]. Genes implicated in pollen-wall development in angiosperms are also present in mosses and lycopsids, suggesting that they may be involved in spore-wall development in basal plants. The molecular genetics of spore/pollen development may thus be highly conserved, despite the large morphological, functional, and developmental differences between spores and pollen. The key innovation was the ability to generate sporopollenin, an extremely tough and resistant polymer protecting haploid spores [3]. The composition of the pollen and pollen tube wall is unique [4] and has varied physically and chemically in different seed plant lineages as well as in response to the environmental aspects of pollination [5]. Studies of numerous mutations affecting pollen wall synthesis [6–18] revealed the importance of pollen wall for the proper function of male gametophyte and the complex regulation of cell wall synthesis by sporophytic and gametophytic tissues [6, 11, 18]. Pollen walls begin to organise before meiosis when meiocytes become surrounded by callose secreted by a tapetum [19].
Callose is a polysaccharide (β-1,3-glucan) that plays a fundamental role in angiosperms in many developmental processes including plasmodesmata formation and cytokinesis as well as in plant responses to biotic and abiotic stresses [20]. Callose is also involved in several stages of development of male gametophytes [21]. The role of callose in cell-wall development in other plant groups is less well defined [2]; for example callose is currently believed to be absent in pteridophytes (excluding only one genus, Selaginella). Callose has been identified in some bryophytes [22] around the spore mother cell, but its link to cell-wall development, if any, is not well understood [2]. Callose is generally sparsely produced in plants, representing only 0.3–5% of the total cell-wall content (Arabidopsis and Miscanthus [23]).
Twelve callose synthase (CalS or glucan synthase-like (GSL)) genes have been identified in Arabidopsis thaliana and classified in one gene family [24–26]. Comprehensive view in organ specific callose synthesis was done in Arabidopsis [27] and in Triticum aestivum [28]. Callose synthases are usually classified in two major groups, one contributing to fertility and cell division: CalS1 (GSL6), CalS5 (GSL2), CalS9 (GSL10), CalS10 (GSL8) and CalS11 (GSL1), and the second providing the reinforcement of structural cell walls: CalS3 (GSL12), CalS7 (GSL7), CalS8 (GSL4), CalS12 (GSL5). The function of CalS2 (GSL3), CalS4 (GSL9) and CalS6 (GSL11) is still unknown [25, 29]. All twelve callose synthases were detected during pollen development (eFP Browser [30]), however only CalS5 represented putative pollen-specific protein being highly expressed in anthers including microspores and pollen, and its role in microgametogenesis was not compensated by other pollen-expressed CalS proteins [6]. CalS5 may have evolved as a key enzyme responsible for callose synthesis required for pollen development and pollen-tube growth in higher plants [31].
Five of the 12 callose synthase genes have been reported to have a function during microsporogenesis and microgametogenesis (CalS5, CalS9, CalS10, CalS11, and CalS12; Table 1). Meiocytes in developing Arabidopsis anther locules are temporarily packed prior to meiosis in a layer of callose to prevent cohesion and fusion and its dissolution results in the release of free microspores [32]. Callose deposition continues during the second meiotic cytokinesis and finally encloses individual microspores within tetrads [33]. The separation of tetrads into free microspores involves degeneration of the callose wall by tapetum-secreted 1,3-beta-glucanase (callase) [34], and the callose wall appears to act as a mould wherein the primexine provides a blueprint for the formation of the exine pattern on the mature pollen grain [6, 35]. Microspores are released after the callose wall degradation and undergo critical pollen mitosis I (PMI) to form immature bicellular pollen grains, where generative and vegetative cells are first separated by a callose cell wall [32]. The degradation of callose in tetrads leading to microspore release is critical for the synchronisation of subsequent microspore development, namely PMI, and is ensured by the extracellular secretion of callase to anther locules by the tapetum [36].
Table 1. Summary of callose synthase proteins their function, and location in the Arabidopsis thaliana.
Proteins active in pollen are underlined.
| Gene/Protein | AGI | Function | References |
|---|---|---|---|
| CalS1 (GLS6) | AT1G05570 | forms a complex with a UDP-glucose transferase and is localized at the cell plate during cytokinesis | [21, 26] |
| regulates plasmodesmal permeability under pathogen infection and mechanical stress | [37] | ||
| CalS2 (GLS3) | AT2G31960 | unknown | [21, 25, 29] |
| CalS3 (GLS12) | AT5G13000 | callose accumulation in plasmodesmata | [21, 38] |
| CalS4 (GLS9) | AT5G36870 | expressed in primordia of branching adaxial buds, exact role unknown | [21, 25, 29] |
| CalS5 (GLS2) | AT2G13680 | callose deposition in the wall and callose plugs of germinated pollen tubes | [6, 7, 21, 39] |
| required for exine formation during microsporogenesis and therefore pollen viability | [6, 7, 40] | ||
| prevents pollen degeneration early in development | [7] | ||
| CalS6 (GLS11) | AT3G59100 | unknown | [21, 25, 29, 41] |
| CalS7 (GLS7) | AT1G06490 | required for callose deposition at the sieve plate | [6, 17] |
| CalS8 (GLS4) | AT3G14570 | regulates plasmodesmal permeability under pathogen infection and mechanical stress | [37] |
| CalS9 (GLS10) | AT3G07160 | involved in the entry of microspores into mitosis | [21, 40, 42] |
| important for the microspore asymmetric division | [40] | ||
| CalS10 (GLS8) | AT2G36850 | involved in pollen development, namely in the entry of microspores into mitosis | [21, 40] |
| required for callose biosynthesis at the cell plate | [43] | ||
| involved in stomatal pattering and deposition at the plasmodesmata | [44] | ||
| CalS11 (GLS1) | AT4G04970 | cell plate formation in sporophytic tissues | [10, 21, 25, 34, 45] |
| prevents callose wall degradation in microspores early in development | [34] | ||
| function in formation of callose wall that separates microspores within tetrads | [34] | ||
| CalS12 (GLS5) | AT4G03550 | synthesis of callose wall that separates microspores within tetrads | [34] |
| prevents callose wall degradation in microspores early in development | [21, 34, 45] | ||
| important for exine formation and pollen wall pattering | [34] | ||
| function in cell plate formation in sporophytic tissues | [21] |
CalS5, CalS9, CalS10, CalS11, and CalS12 have been shown to be involved in pollen development at and around the mitotic stage. CalS5 participates in callose deposition in the pollen wall and in the plugs of growing pollen tubes [7]. It is also essential for the formation of the callose wall surrounding pollen mother cells [40]. Three distinct roles have been defined for CalS5 in pollen development: patterning the exine layer, forming callose in pollen tubes, and preventing pollen degeneration early in development [7]. CalS11 and CalS12 play at least partially redundant roles in pollen development [34] but are expressed at lower level than CalS5 [6]. CalS9 and CalS10 are also co-expressed in lower amounts than CalS5 [6]. They are responsible for the formation of the callose wall separating the microspores in tetrads and during pollen-grain maturation. CalS12 plays also an important role in stress-induced callose deposition [46]. CalS9 and CalS10 are independently required for an asymmetric microspore division and entry of microspores into mitosis [40]. CalS9 is required for the entry of microspores into the first and second mitotic divisions and may function in later pollen development after the free-microspore stage [42].
There are two types of callose, peripheral callose and interstitial callose, which are synthesised in the tetrad by different callose synthases. The peripheral callose of the tetrad is absent in the knockout mutant cals5-2, but the interstitial callose is present. This observation clearly indicates that CalS5 is responsible for the synthesis of peripheral callose [6]. Moreover, the main regulator of auxin signalling, auxin response factor 17 (ARF17), directly binds to the CalS5 promoter to regulate its expression for callose synthesis [47]. CalS11 and CalS12 genes are responsible for the interstitial tetrad callose synthesis [34].
The evolutionary relationships amongst the orthologues of plant callose synthases in model plants are poorly known [21, 48, 39]. Here we analysed a large data set of 1211 amino acid sequences of callose synthase family members to classify all representatives available across angiosperms, gymnosperms, lycopods, and bryophytes to determine the associated evolutionary processes in the plant kingdom. We demonstrated the expression of callose synthase family members in A. thaliana and Nicotiana tabacum, with special emphasis on different pollen developmental stages.
Materials and methods
A. thaliana and N. tabacum gene expression data
The expression data for A. thaliana were obtained from eFP Browser [30] and from other publicly available resources as described previously [49, 50, 51]. Briefly, the transcriptome datasets were downloaded from the NASCArray microarray database through the AffyWatch service [52] and normalized using freely available dChip 1.3 software (http://www.dchip.org) to the median probe intensity level; model-based gene-expression values were based on the Perfect Match-only model [53, 54].
Total RNA microarray data for N. tabacum callose synthases were obtained from previously published datasets [55, 56] and [57]. Subcellular fractions related to callose synthases mRNA translation and storage (polysomes and RNA storage EPP particles [58]) were isolated by sucrose gradient centrifugation as described previously [59]. From both fractions, total RNA was isolated using RNeasy Plant Mini Kit (Qiagen) and used for the hybridization of Agilent 44K Tobacco Gene Expression Microarray as described in [59]. Again, the transcriptomic datasets were normalized using publicly available dChip 1.3 software and further analysed as described above and in [57].
Searching transcriptomes and genomes for CalS homologues
CalS homologues were identified by BLASTP searches using A. thaliana CalS1-12 proteins from the TAIR database (https://www.arabidopsis.org/; Table 1) to query NCBI protein databases (http://www.ncbi.nlm.nih.gov/). The BLASTP searches used default parameters, adjusted to the lowest E-value. The duplicates from all searches were eliminated. The Phytozome version 11 database (https://phytozome.jgi.doe.gov) was next searched for callose synthase proteins not found by BLASTP. Finally, we conducted an iterative search of the UniProt database (http://www.uniprot.org/). We analysed all sequences independently of their annotations, with no prior assumptions.
Sequence alignment
Amino acid sequences were aligned using the Clustal Omega algorithm [60] in the Mobyle platform [61], with homology detection by HMM–HMM comparisons [62]. Protein isoforms with the same length were also used, because the differential expression patterns producing protein isoforms from various tissues suggested that isoforms could have different biological functions in vivo [63].
Phylogenetic reconstruction
Maximum likelihood (ML) topology searches were performed with MEGA 7 [64] based on the JTT matrix-based model. Initial tree(s) for the heuristic search were obtained by applying the neighbour-joining and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model and then selecting the topology with the best log likelihood value. A discrete gamma distribution was used to model differences in evolutionary rate amongst sites (five categories (+G, parameter = 0.9166)). The model of rate variation allowed some sites to be evolutionarily invariable ([+I], 0.2289% sites). The analyses included 1211 (ML analysis) and 655 (ML and rooted MP analyses) amino acid sequences. The final data set contained a total of 4150 and 3435 positions, respectively. Phylogenetic trees were constructed and modified with iTOL v3.4 [65]. A bootstrap analysis under the ML criterion was run in RAxML due to the size of the data set [66]. The analyses were run with optimised equilibrium frequencies using the GTR model of evolution for tree inference.
Maximum-parsimony (MP) analysis was conducted in MEGA 7 using the Tree-Bisection-Regrafting algorithm with the initial trees obtained by the random addition of sequences. The analysis included 655 amino acid sequences. The final data set contained a total of 4970 positions.
Timetrees for each callose synthase with function in pollen were computed in MEGA 7 [64] using estimated divergence times for all branching points in a tree applying an approach based on the relative-time (RelTime) method [67], which does not require assumptions for variations in lineage rate. Divergence times for all branching points in the topology were calculated using ML based on the JTT matrix-based model.
Identification of domains and conserved amino acid sequence motifs
The Pfam Motif Library [68], NCBI DART [69], and MEME 4.11.2 [70] were used to analyse the conserved motifs of the selected proteins and the reference sequences. The MEME search was set to identify a maximum of 50 motifs for each protein with a wide sequence motif from 2 to 50 and total number of sites from 2 to 600. The number and arrangement of introns and exons were analysed using Gene Structure Display Server version 2.0 [71] by aligning the coding sequences with the genomic sequences.
Results
Analysis of 12 CalS homologues in A. thaliana
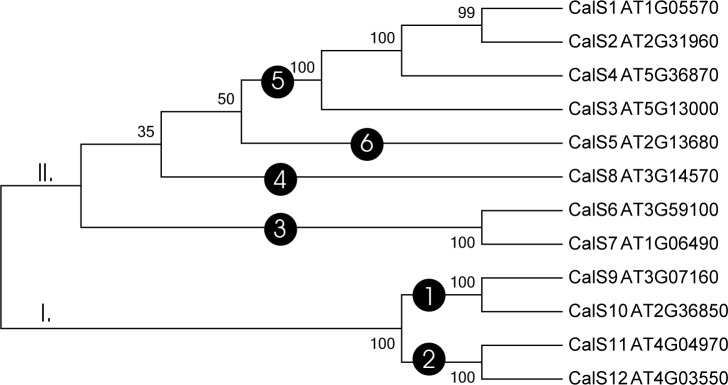
The analysis of the 12 CalS homologues produced a phylogenetic tree depicting the relationships amongst all callose synthase family members in A. thaliana (Fig 1). The tree had two main groups, one containing CalS1-CalS8 and the second containing CalS9-CalS12. Subfamilies forming separate clades were CalS9/10 in clade 1, CalS11/12 in clade 2, CalS6 and CalS7 in clade 3, CalS8 in clade 4, CalS1, CalS2, CalS3 and CalS4 in clade 5, CalS5 in clade 6.
Fig 1. Phylogenetic tree of 12 Arabidopsis thaliana callose synthases.
The evolutionary history was inferred using maximum likelihood. The bootstrap support values are shown next to the branches. Accessions are listed in Table 1.
Expression of callose synthase genes in different tissues of A. thaliana and in the pollen stages of N. tabacum
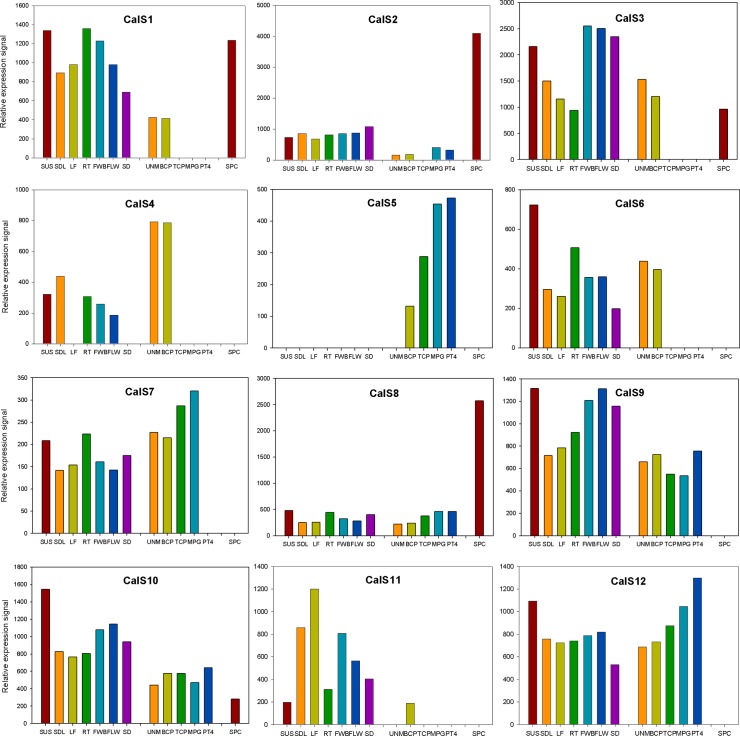
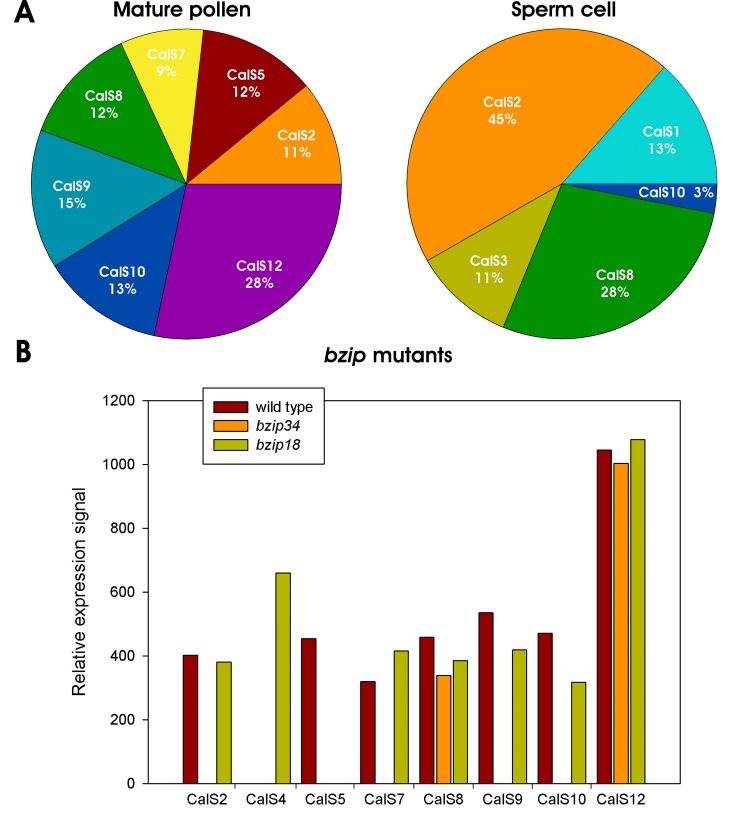
The callose synthase expression levels in different plant tissues, including male gametophyte, are shown in Fig 2. All callose synthase genes, except CalS5 were expressed in most sporophytic tissues at various level. Similarly, detectable concentrations of all 12 callose synthases were found in at least several stages of A. thaliana pollen development [30, 49, 72]. However, pollen-expressed CalS genes showed three distinct expression patterns. The first group contained genes expressed only early in male gametophyte development that later disappeared or were expressed at very low level (CalS1, CalS2, CalS3, CalS4 and CalS6). Based solely on the expression profile in the male gametophyte, CalS11 can be put into this group too, however other characteristics (see below) associates this gene more likely with the group two. The second group comprised callose synthases that were expressed throughout male gametophyte development at higher abundance, often being accumulated in later developmental stages (CalS7, CalS8, CalS9, CalS10 and CalS12). All these proteins were expressed also in sporophytic tissues. The last expression group contained only one member, CalS5, that also accumulated throughout pollen development and progamic phase but unlike others, was pollen-specific. Finally, specific set of genes, partially overlapping with groups one and two, then comprised callose synthases that were active in male gametes, sperm cells (CalS1, CalS2, CalS3, CalS8 and CalS10). Although all these genes were expressed also in the sporophyte, they differed in their expression patterns throughout pollen development (Fig 2). Interestingly, sperm-cell expression of most of them was rather strong. Taken together, the most abundantly expressed callose synthase proteins in A. thaliana mature pollen were CalS12, CalS9 and CalS10, whereas sperm cells expressed mostly CalS2, CalS8 and CalS1 (Fig 3A).
Fig 2. Expression profiles of 12 callose synthases in different sporophytic tissues and during male gametophyte development of Arabidopsis thaliana.
SUS, suspension cultures; SDL, seedling; LF, leaf; RT, root; FWB, flower buds; FLW, flower; SD, seed; UNM, uninuclear microspore; BCP, bicellular pollen; TCP, tricellular pollen; MPG, mature pollen grain; PT4, 4h pollen tubes; SPC, sperm cells.
Fig 3. Callose synthase expression in Arabidopsis thaliana pollen.
A) Abundance of expressed CalS transcripts in mature pollen and sperm cells. B) Expression profiles of pollen-expressed CalS transcripts in wild type Col-0 and bzip18/- and bzip34/- mutant A. thaliana mature pollen.
bZIP family transcription factors bZIP18 and bZIP34 were previously shown to be involved also in the regulation of pollen wall formation [11, 18]. Transcriptomic analyses of pollen deficient in these proteins showed that many mature pollen-expressed callose synthases are in the regulons of either bZIP18, bZIP34 or both. Of seven callose synthases expressed in mature pollen, only two (CalS8 and CalS12) were not affected in bzip18 or bzip34 mutants (Fig 3B) suggesting that the regulation of their expression was independent of the activity of studied bZIP proteins. On the contrary, CalS2, CalS7, CalS9 and CalS10 expression was strongly downregulated in bzip34 mutant pollen. The most severely affected was pollen-specific gene CalS5, that was downregulated in both bzip18 and bzip34 mutants independently. Finally, CalS4 that is usually expressed only during early male gametophyte development, was activated also in mature pollen in bzip18 mutant pollen. Altogether, our data suggest that pollen callose synthases may be at least partly regulated by bZIP family transcription factors together with other proteins involved in the synthesis of cell wall formation [11, 18].
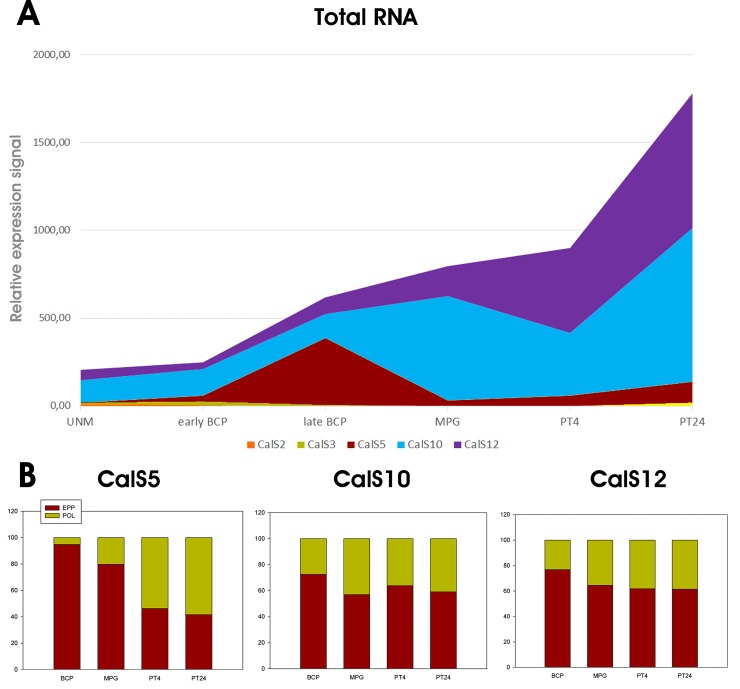
Eight callose synthase subfamilies were found to be encoded in N. tabacum (CalS2, CalS3, CalS5, CalS7, CalS8, CalS10, CalS11 and CalS12). Of them, only five were expressed during pollen development (Fig 4), whereas remaining three were much more abundant. CalS5 abundance peaked at late bicellular stage before pollen maturation, whereas CalS10 and CalS12 were most strongly expressed after pollen germination, in growing pollen tubes (Fig 4A). Interestingly, the overall quantity of transcripts of three most abundant callose synthase transcripts was not reflected by their transitional efficiency [58, 73]. Whereas CalS10 and CalS12, peaking during progamic phase, were similarly distributed between storage EPP complexes and actively translating polysomes throughout the whole pollen development, CalS5 was more intensely translated after pollen germination when more callose synthase activity was also needed for pollen tube wall synthesis (Fig 4B).
Fig 4. Callose synthase expression profiles in Nicotiana tabacum male gametophyte.
A) Expression profile of five male gametophyte-expressed CalS mRNAs during pollen development and progamic phase, B) Distribution of transcripts encoding three most abundant tobacco pollen CalS mRNAs (CalS5, CalS10, CalS12) between actively translated polysomal and storage EPP fractions. UNM, uninuclear microspore; early BCP, early bicellular pollen; late BCP, late bicellular pollen; MPG, mature pollen grain; PT4, 4h pollen tubes; PT24, 24h pollen tubes; EPP, mRNA storage EPP complexes; POL, polysomal fraction.
Identification and phylogenetic analysis of the CalS family in plants
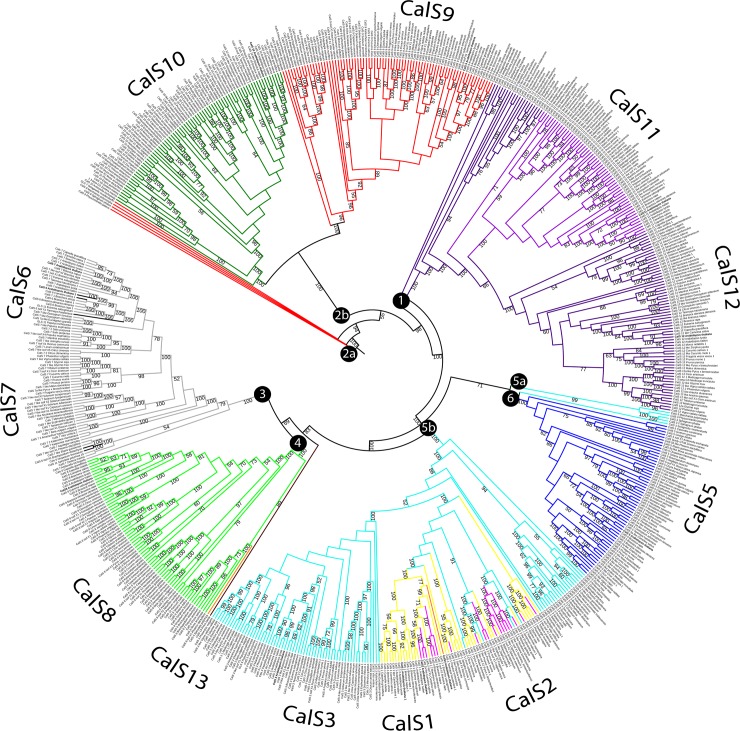
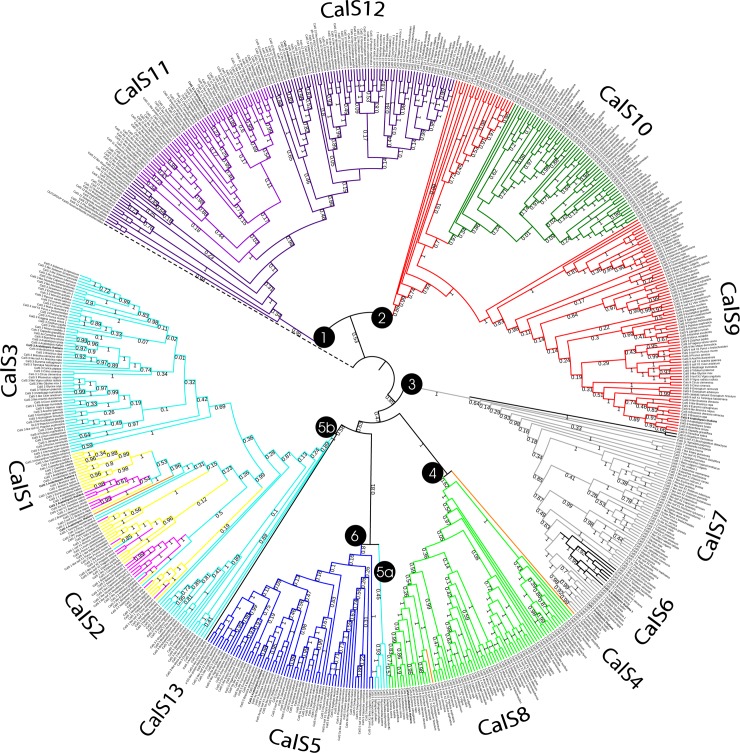
The 1211 sequences assembled from 101 plant species were analysed for the presence of CalS and CalS-like proteins. The evolutionary relationships amongst the callose synthases were determined using ML and MP analyses based on multiple alignments of the CalS proteins. The evolutionary hypotheses from these analyses were highly congruent. Most of the CalS proteins in the phylogenetic tree containing 1211 protein sequences formed main clades corresponding to the distribution of the 12 family members previously found in A. thaliana (Fig 1 and S1 Fig). Not all species contained orthologues of all 12 proteins; the number varied from four to twelve. Only a few species contained more than single-copy paralogues of a particular protein. Five CalS proteins directly linked to pollen function evolved twice; CalS9, 10, 11, and 12 formed a well-defined clade (clades 1 and 2), and the most prominent pollen-specific protein, CalS5 (clade 6), was embedded inside the subfamilies in the MP tree that were expressed in pollen at lower lever (Figs 5 and 6 and S1 Fig). Therefore, CalS5 formed a sister group to the pollen non specifically-expressed group comprising CalS3/2/1, CalS8, CalS7, and CalS6 in the ML tree (clades 3, 4 and 5; Fig 5 and S1 Fig). Only three subfamilies, CalS5, CalS9, and CalS11, however, formed monophyletic, conserved clades. Similar situation was found for other model plant with sequenced genome, Nicotiana tabacum, allotetraploid species that arose from the hybridization of N. sylvestris and N. tomentosiformis [74]. Only nine CalS proteins were found in Nicotiana, CalS1 from clade 5, CalS4 (clade 4) and CalS6 (clade 6) are missing. Members from each phylogenetic group (order) tended to cluster together within a given subfamily represented by a clade, suggesting that different callose synthase subfamilies could have expanded after the divergence from their common ancestor to form their own subclasses within each CalS subfamily (e.g. bryophytes, lycophytes, monocots, and dicots).
Fig 5. Unrooted phylogenetic tree of 655 proteins sequences of the callose synthase.
The evolutionary history was inferred using maximum likelihood. Bootstrap support values ≥50% are shown on the branch above. The ML log likelihood -482640.553247. The analysis included 655 amino acid sequences and 3435 positions in the final data set. Numbers in circles indicate main clades described in the text.
Fig 6. Callose synthase phylogeny in terrestrial plants inferred using maximum parsimony.
The tree is rooted by green alga Auxenochlorella protothecoides representing one of possible outgroup for terrestrial plants. The most parsimonious tree with length 90012, CI = 0,223978, RI = 0,828990 is shown. The data contains 655 protein sequences and 4970 positions in the final dataset. Bootstrap support values ≥50% are shown next to the branches. Numbers in circles indicate main clades described in the text.
CalS9 formed two distinct clades (Figs 5 and 6 and S1 Fig), the first formed in angiosperms and the second grouping bryophytes (Physcomitrella and Sphagnum) and lycopods (Selaginella). CalS10 formed monophyletic clade inside CalS9. The CalS11 clade was monophyletic but embedded inside CalS12. CalS6 formed a separate clade mixed with CalS7 (clade 3). The CalS7 subfamily contained the basal angiosperm Amborella trichopoda in the basal part of the clade. CalS3 was divided into three groups, one found in bryophytes (Physcomitrella and Sphagnum) and lycopods (Selaginella) in the clade 5a and basal for CalS5, one present mostly monocots and basal for CalS1 and CalS2, and the third CalS3 clade grouping only dicots, which clustered together with the second group. The CalS8 subfamily formed a separate clade 4, but contained Spirodela polyrhiza CalS4 as a sister group and embedded Theobroma cacao CalS4 inside the CalS8 subfamily.
Only one representative from gymnosperms, the CalS13 protein from Pinus taeda, was supported in a basal position of the CalS3 clade. The sequence from P. taeda was used, although it was about 50% shorter than the sequences in the rest of the matrix, because only the most informative sites were included. Basal angiosperms from the study [39] were similarly retained in the analyses.
We next determined the divergence of the time estimates for all branching points in the callose synthase trees associated with pollen development. Relative-timed ML phylogenetic trees established for the callose synthase subfamilies involving pollen (CalS5, CalS9, CalS10, CalS11, and CalS12) strongly supported closer relationships within the main taxonomic groups (orders, S2–S6 Figs). Branch lengths indicated the length of time between nodes on a relative timescale. Relative time within each branch did not differ substantially, indicating a common origin and higher conservation in the callose synthase proteins. Only the relative times for bryophytes and lycopods diverged greatly, similarly to the evolutionary distance from angiosperms (S3 and S6 Figs). These results suggested relatively short evolutionary time of divergence within each subfamily.
The distribution of conserved amino acid domains and motifs within the CalS clades
The comparison of the sequences of the callose synthase subfamilies indicated that members of the same subfamily were closely related to each other and distantly related to the members of other subfamilies, consistent with the results of the phylogenetic analyses. The 12 CalS genes in A. thaliana were distributed over the five chromosomes [26]: chromosome 1, GLS6 (CalS1) and GLS7 (CalS7); chromosome 2, GLS2 (CalS5), GLS3 (CalS2), GLS8 (CalS10) and GLS11 (CalS6); chromosome 3, GLS4 (CalS8) and GLS10 (CalS9); chromosome 4, GLS1 (CalS11) and GLS9 (CalS4), GLS5 (CalS12); and chromosome 5, Chromosome 5 and GLS12 (CalS3). The genes expressed abundantly in pollen were on chromosomes 3–5 (Table 1).
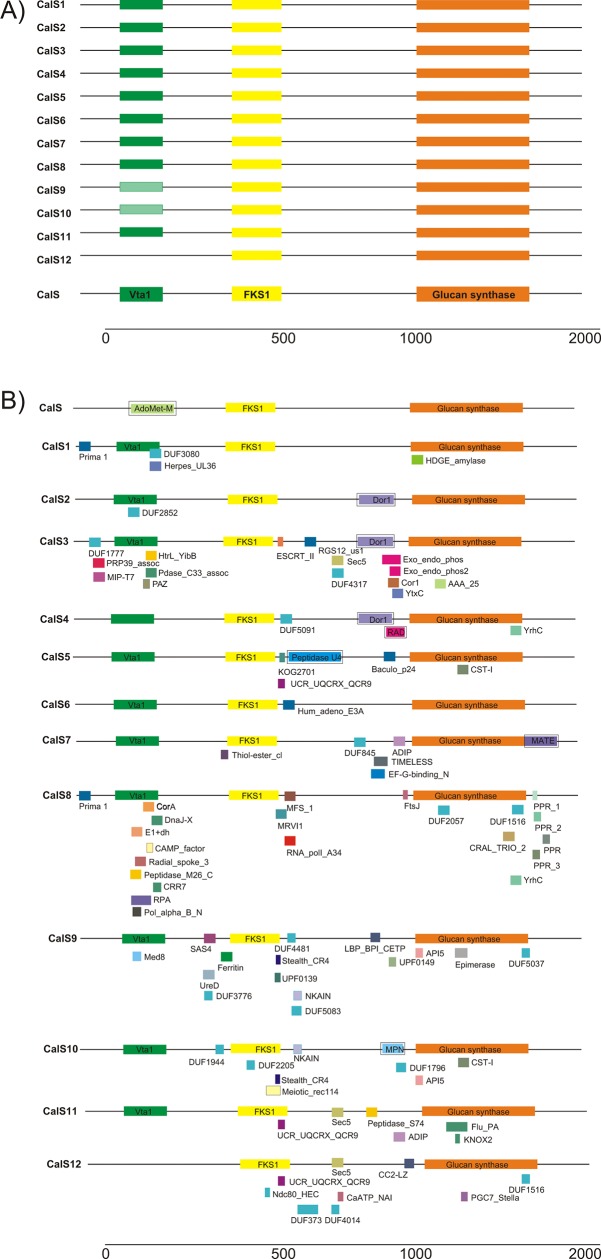
Most of the CalS genes encoded proteins of about 2000 amino acids in length. CalS proteins typically consisted of three conserved domains: Vta1, FKS1, and glucan synthase (Fig 7). The Vta1 domain was present in all subfamilies but sometimes was not found in CalS9 and Cals10 proteins. Vta1 is involved in the transport of the multivesicular bodies, an endosomal compartment involved in sorting membrane proteins for degradation in lysosomes [75]. This division fully corresponded to the distribution of the CalS subfamilies, where CalS9 and Cals10 formed one well-supported clade (Figs 4 and 5). The FKS1 domain was highly conserved across the CalS proteins of terrestrial plants and was homologous with fungal FKS genes encoding an integral membrane protein, a subunit of 1,3-beta-D-glucan synthase [26, 76]. Further comparative analysis of CalS proteins in Pfam identified 81 types of protein domains (Fig 7). Some of them were unique for particular subfamilies (e.g. DUF3080 in CalS1 or KNOX2 in CalS11), and some were shared amongst two or more subfamilies. For example, UCR_UQCRX_QCR9 (ubiquinol-cytochrome C reductase) domains were shared amongst CalS5, CalS11, and CalS12, whereas API5 (apoptosis inhibitory protein) was shared by CalS9 and CalS10, and NKAIN (Na, K-ATPase interacting protein) was found in CalS9 and CalS11. S1 Table provides detailed information of the type, hypothetic function, and distribution of each domain within the species. Most of the domains were found in CalS8 (22) and CalS3 (16), and only one was found in CalS6. Callose synthases involved in the development of male gametophytes usually contained fewer domains (five in CalS5, six in CalS11, and nine in CalS10 and CalS12). To sum up, we found highly variable multidomain structure of callose synthase proteins within plants. It is generally believed that multidomain proteins evolved under selective pressure during evolution to create new function. From this point of view, CalS8, CalS3 and CalS9 seem to be under the highest evolutionary pressure. Moreover, in different phylogenetic groups of plants different domains evolved in response to different physiological and developmental signals. The detection of the MEME motifs indicated other structural diversification of the callose synthase proteins. MEME works by searching for repeated, ungapped sequence patterns that occur in the protein sequences. A set of proteins that interact with a single host protein may do so via similar domains. The details of the 50 putative motifs identified are shown in S2 Table. 44 motifs were conserved across all callose synthase subfamilies, and six motifs were more specific for two or more subfamilies. CalS1/2/3 and CalS5 contained motif 42, corresponding to clades 5 and 6 (Figs 5 and 6). CalS9/10 contained motif 50 that was specific to clade 6. Interestingly, CalS8/11/12 did not contain motif 14, Cal6/8 did not contain motif 23, CalS6/8/10/11/12 did not contain motif 32, and CalS6/10/11/12 did not contain motif 47. The functions of each motif were identified by searching Pfam, showing that nine motifs encode glucan synthase domain, two motifs belong to the FKS1 domain, five motifs belong to transmembrane domains, while the remaining 34 motifs do not appear to be associated with any domain.
Fig 7. Consensus domains in the callose synthase proteins.
A) Three main domains from the Vta1 superfamily, 1,3-beta-glucan synthase subunit FKS1, and the 1,3-beta-glucan synthase superfamily. B) Specific domains for each callose synthase. Domains were predicted by Pfam, and the domain in the boxes was predicted by DART. See S1 and S2 Tables for details.
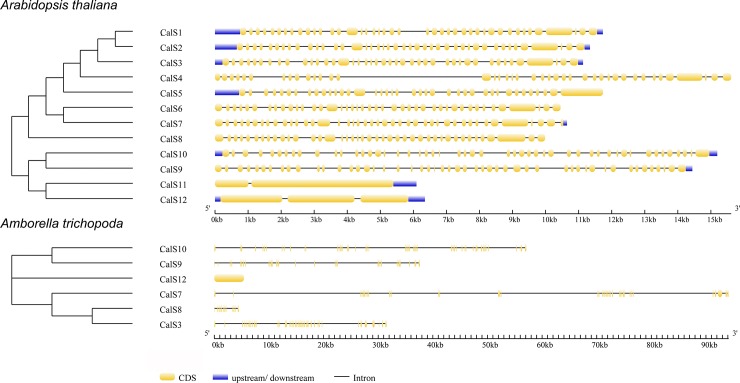
Callose synthase genes showed remarkably complex exon/intron structure and with the exception of CalS11 and CalS12, they usually contained over 40 exons (Fig 8). For example, CalS1 gene comprised 42 exons and 41 introns and was transcribed into a 6-kb mRNA [25]. Similar complex arrangement was found also basal angiosperm (A. trichopoda) that highlighted that this feature is characteristic for all angiosperm phylogenetic group (Fig 8).
Fig 8. Comparison of the number and arrangement of introns and exons for representative species from a derived angiosperm (Arabidopsis thaliana) and a basal angiosperm (Amborella trichopoda).
Discussion
Expression patterns of members of the callose synthase family
Genome analyses indicated that A. thaliana contained all 12 members of the callose synthase family. N. tabacum, however, contained only eight callose synthases, and three callose synthases abundantly expressed in pollen were distributed significantly differently during pollen development. CalS9, CalS10 and CalS12 were expressed at similar or higher levels than CalS5 in A. thaliana, although the non-redundant function for male gametophyte development was demonstrated only for pollen-specific CalS5 [6]. However, CalS5 expression in N. tabacum was lower than that of CalS10 and CalS12 in total pollen and this trend was even more apparent during the progamic phase. On the contrary, the putative translation activity of CalS5 transcript was higher than that of CalS10 and CalS12 in growing pollen tubes (Fig 4), This gradual translational activation in growing pollen tubes showed similar pattern as the translation of pollen-specific gene ntp303 encoding pollen tube-wall glycoprotein p69 [58, 73].
CalS9/CalS10 and CalS11/CalS12 were closely related. The CalS10 clade was embedded within CalS9 in the phylogenetic tree, and CalS11 was similarly embedded within the CalS12 clade. The above results supported the suggestion in [42] that these members of the CalS family acted in a complex, or more likely, that these two pairs played at least partially redundant roles in pollen development [34]. In Arabidopsis, such hypothesis was supported only for co-expressed proteins CalS9 and CalS10, whereas the expression profiles of CalS11 and CalS12 were different; CalS12 was the most abundant late pollen-expressed callose synthase but CalS11 expression was much lower and at earlier developmental stages. It seemed that in N. tabacum pollen development, only CalS10 and CalS12 from these pairs were active. CalS10 and CalS12 may have specific developmental tasks in species with bicellular pollen, such as N. tabacum. Both members of these pairs, though, still may act in pollen development in species shedding tricellular pollen, such as A. thaliana. We can thus speculate which state is ancestral and which derived. In [77] proposed that bicellular pollen evolved secondarily from tricellular ancestors during shifts away from a rapid life cycle or from limited reproduction. We can follow up this hypothesis that during the evolution of derived species with bicellular pollen, reduction of possible redundant callose family members occurred and their role in pollen development was played by only one partner, i.e. CalS10 and CalS12 in this case. Interestingly, genes encoding eight CalS family members were identified in tobacco genome, however not all of them were expressed. Therefore, they might represent pseudogenes or might be silenced by the activity of matching miRNAs.
The absence of CalS6, CalS7 and CalS8 in tobacco pollen is similar to findings in [26]. They speculated that the absence of these callose synthases in EST database (based on leaf samples) suggested that these genes might be expressed at very low levels or that their expression pattern might be induced special conditions such as pathogen infection. However, since we found pollen-abundant CalS5 expressed also in N. tabacum [57]; we did not identify CalS6 and we can also assume that the expression of CalS7 and CalS8 is very limited in wild type pollen.
Phylogenetic reconstruction and evolutionary history of CalS in terrestrial plants
Callose is synthesised by callose synthases encoded by members of the CalS gene family, which vary amongst terrestrial plants. A. thaliana contains 12 members in two main clades [21]. The number of CalS orthologues, however, varies within individual groups of terrestrial plants. Callose synthase subfamilies are structurally similar and likely diverged from a common ancestral glucan synthase domain, which is conserved in all subfamilies. Multiple CalS genes may have evolved in higher plants to catalyse callose synthesis in different locations and in response to different physiological and developmental signals [26]. Our phylogenetic analyses were designed to identify the relationships within the callose synthases subfamilies in detail.
In contrast to [6], we found two main groups and six subgroups in the phylogenetic tree of A. thaliana callose synthases. In [21] described four main subfamilies but did not include the support values for the tree branches. They also reported only a composite tree using ClustalW2 for phylogenetic analysis. According [21], the first subfamily contained CalS11 (AtGSL1), CalS12 (AtGSL5), CalS10 (AtGSL8), and CalS9 (AtGSL10), the second subfamily contained CalS5 (AtGSL2), CalS2 (AtGSL3), CalS1 (AtGSL6), and CalS3 (AtGSL12), the third subfamily comprised CalS7 (AtGSL7) and CalS6 (AtGSL11), and the fourth subfamily contained CalS8 (AtGSL4). The descriptions of the subfamilies, however, is unusual and lacking appropriate experimental support, and CalS4 (AtGLS9) was not included in any group. We analysed the same set of samples (Fig 1), generated an ML tree, and proposed the same principal two groups containing six subfamilies: subfamily 1, CalS9 and CalS10; subfamily 2, CalS11 and CalS12; subfamily 3, CalS6 and CalS7; subfamily 4, CalS8; subfamily 5, CalS1, CalS2 and CalS3; and subfamily 6, CalS5.
Our comprehensive phylogenetic analysis of the CalS family included 1211 and 655 sequences, respectively (Figs 5 and 6 and S1 Fig) from 101 plant species. The sequences grouped into six distinct clades in the phylogenetic tree (S1 Fig). Early events in the diversification of embryophytes gave rise to mosses, liverworts, and hornworts. All bryophyte lineages share a life cycle in which the gametophyte (haploid phase) dominates, with a sporophyte (diploid phase) dependent on the maternal gametophyte. Vascular plants instead have a dominant sporophytic phase and more or less reduced gametophytes. A basal branch of bryophytes in the plant phylogenetic tree would support the hypothesis that the dominant life cycle of gametophytes is plesiomorphic in embryophytes [78]. The proteins appeared to be conserved, and in a few cases seemed to diversify after the divergence of bryophytes and vascular plants, despite differences in spore/pollen development. The evolutionary history of the callose synthase subfamilies supports this hypothesis. CalS9 and CalS10 are clearly genetically linked, with both subfamilies highly supported (86% bootstrap support (BS), Fig 6), but CalS9 formed a second basal clade containing bryophyte (Physcomitrella and Sphagnum) and lycopod (Selaginella) callose synthases. CalS12 and CalS11, respectively, had a similar pattern. Moreover, CalS10 and CalS11 were not identified in any bryophyte or lycophyte, respectively, so they may have diverged by a duplication event in vascular plants after the divergence of bryophytes and lycopods. The results also supported the putative basal position of Selaginellales relative to all other vascular plants [78], because all members of the callose synthase subfamilies clustered together with bryophytes.
The only gymnosperm representative, Pinus taeda, contained the CalS13 protein. It was supported in the basal position of the CalS3 clade but only with low statistical support (in ML tree 69% BS and in MP tree 58% BS, see Figs 5 and 6). Pollen-wall structure and development differs greatly between gymnosperms and angiosperms [2]. Callose walls in gymnosperms form around the pollen mother cells and subsequently extend around each pollen microspore [2]. There, CalS13 becomes strongly expressed in the aperture region surrounding the emerging pollen tube after germination. In gymnosperms pollen tube cell wall does not contain callose and, accordingly, CalS13 does not extend into the pollen tube [19, 39].
The phylogenetic analyses identified six main highly supported clades containing all callose synthase subfamilies. However, group 2 and 5 were divided into two clades, first in ML analysis (S1 Fig) and the second in both, ML and MP (Figs 5 and 6). Clades 1, 2, and 6 in the trees (Figs 5 and 6) contained callose synthase subfamilies involved in late pollen development and pollen tube growth. Clade 1 contained CalS11 and CalS12 (100% BS), and clade 2 contained CalS9 and CalS10 (100% BS). CalS6 and CalS7 were in clade 3 (100% BS), CalS8 was in clade 4 (100% BS, Fig 5), and CalS1, CalS2, and CalS3 were in clade 5 (100% BS). Clade 6 contained CalS5 (100% BS, Fig 5). The roles of most of these subfamilies are not yet fully understood. CalS1 functions in cell-plate formation in sporophytic tissues [21], and CalS7 is required for the deposition of callose at the sieve plate [31]. All three clades formed distinct subclades with high support (99–100% BS, Fig 6). CalS6 and CalS4 may have evolved by duplication from CalS7 and CalS8, respectively. CalS2/1 in clade 5b may have evolved from CalS3 after the diversification of bryophyte CalS3 (clade 5a), which is also supported by the wider distribution of CalS3 than CalS2 and CalS1. In the light of CalS9/10 and CalS11/12 position in phylogenetic trees and their possible redundant function, we can agree with [11] that CalS1 and CalS2 are also possibly redundant. Observed high sequence homology also supports this statement.
Our data are in agreement with the report in [42] that most closely related CalS family members may have similar functions as a complex responsible for callose synthesis. We focused our attention mainly on five subfamilies of the 12 callose synthases that function during microsporogenesis and microgametogenesis (CalS5, CalS9, CalS10, CalS11 and CalS12). CalS9 and CalS10 were closely related within the CalS family. The CalS10 clade was embedded within CalS9 in the phylogenetic tree (97% BS in MP tree, Fig 6), supporting the suggestion in [42] that these members of the CalS family work as a complex. In [40], however, found that CalS10 and CalS9 were independently required for the development of male gametophytes and plant growth. In [79] reported similar results for cellulose synthase complexes that contained three cellulose synthase homologues. CalS11 and CalS12, which form a separate clade (95% BS in MP tree, Fig 6), are also genetically linked and have redundant roles in pollen development and fertility, especially in the formation of walls separating the microspores of the tetrad and late in the maturation of pollen [34].
The CalS5 subfamily formed a separate clade (and 100% in ML tree and 80% BS in MP tree, Figs 5 and 6), but its position was not clear. We hypothesized that all callose synthase subfamilies that play a role in the development of male gametophytes evolved separately from other CalS families. This hypothesis was supported by the ML analysis (Fig 5), where the clade expressing CalS5 in pollen was highly supported (100% BS). The monophyletic position of this group, however, was violated by bryophyte (Marchantia polymorpha, Physcomitrella patens, and Sphagnum falax) and lycopod (Selaginella) members of the CalS3 subfamily, which clustered together in clade 5a (Figs 5 and 6). Moreover, CalS5 in the more complete ML tree (S1 Fig) and rooted MP tree (Fig 6) was embedded in the subfamilies that were not expressed in mature pollen but were expressed in early stages of male gametophyte development instead (CalS1, CalS2, CalS3 and CalS4, clade 5). Some positions of CalS5 in basal angiosperms remained problematic, because only partial sequences were available (but formed a separate clade with unresolved positions from each other).
Interestingly, most callose synthases expressed in A. thaliana sperm cells were phylogenetically related to CalS5, forming clades 4 a 5 (CalS1, CalS2, CalS3 and CalS8), although none of them was apparently pollen-specific. The only exception was CalS10 which also showed expression signal in sperm cells but its expression was much lower than that of the other sperm cell-expressed genes. It would be interesting to investigate whether the sequence diversification between clades 5 (CalS1/2/3) and 6 (CalS5), that was also highlighted by the expression profiles of their members, reflects only regulatory or also functional specialization.
Our analyses led to the description of highly conserved Vta1-FKS1-GluSynt module (Fig 7) of callose synthases in all subfamilies across the plant kingdom. The N-terminal Vta1 domain however, was sometimes missing in CalS9 and CalS10. This division was fully consistent with a close relationship between these two subfamilies (Figs 5 and 6). In CalS9, the Vta1 was missing in most of the bryophytes, except for M. polymorpha, basal angiosperms, and monocots. Vta1 in CalS9 was also absent in dicots, except for the genera Arabidopsis, Brassica and Eutrema. Vta1 in CalS10 was absent from most of the plant species, except for the monocot Zostera marina (Alismatales) and the dicots Medicago truncatula (Fabales) and Nicotiana alata (Solanales). These findings suggested the convergence amongst individual plant species. In contrast, the evolution of other regions of the proteins was more dynamic. The comparative analysis of the CalS proteins identified 81 types of protein domains (Fig 7). Seven subfamilies shared more variable C termini (CalS4, 5, 6, 7, 10, 11, and 12), but others varied across the entire region (CalS1, 3, 8, and 9). Bryophytes and basal angiosperms more often contained motifs not shared with other plants, in agreement with their phylogenetic positions.
The 12 callose synthase genes can be divided into two groups based on gene structure in Arabidopsis thaliana [26]. The first group harbours CalS11 and Cals12 genes which have only two or three exons and the second group contains from 40 to 50 exons in CalS1- CalS10 (Fig 8). Nevertheless, this composition is not same in all plant species as we shown in basal angiosperm Amborella trichopoda, where e.g. CalS12 contains only one exon and CalS3 43 exons. We can point out that the structural divergence among different callose synthases within plant kingdom is generally high and clearly corresponds with their phylogenetic positions of studied species.
Supporting information
Highlighted domains are found in different plants.
(XLSX)
Sequence logos indicates main 50 conserved sequence patterns.
(DOCX)
The evolutionary history was inferred by using maximum likelihood; the tree with the highest log likelihood (-979085.0765) is shown. The analysis included a total of 4150 positions in the final data set. Main branches with circles are described in the text.
(TIF)
The timetree shown was generated using RelTime. The estimated log likelihood value of the topology shown is -57276.0114. A discrete gamma distribution was used to model the differences in evolutionary rate amongst sites (five categories (+G, parameter = 0.5505)). The model of rate variation allowed some sites to be evolutionarily invariable ([+I], 11.2557% sites). The tree is drawn to scale, with branch lengths measured in relative number of substitutions per site. The analysis included 105 amino acid sequences, and the final data set had a total of 2190 positions.
(TIF)
The timetree shown was generated using RelTime. The estimated log likelihood value of the topology shown is -91929.9006. A discrete gamma distribution was used to model the differences in evolutionary rate amongst sites (five categories (+G, parameter = 0.8543)). The model of rate variation allowed some sites to be evolutionarily invariable ([+I], 7.5727% sites). The tree is drawn to scale, with branch lengths measured in relative number of substitutions per site. The analysis included 127 amino acid sequences, and the final data set had a total of 2476 positions.
(TIF)
The timetree shown was generated using RelTime. The estimated log likelihood value of the topology shown is -54519.4579. A discrete gamma distribution was used to model the differences in evolutionary rate amongst sites five categories (+G, parameter = 0.5253)). The model of rate variation allowed some sites to be evolutionarily invariable ([+I], 0.7256% sites). The tree is drawn to scale, with branch lengths measured in relative number of substitutions per site. The analysis included 104 amino acid sequences, and the final data set had a total of 2274 positions.
(TIF)
The timetree shown was generated using RelTime. The estimated log likelihood value of the topology shown is -54506.2366. A discrete gamma distribution was used to model the differences in evolutionary rate amongst sites (five categories (+G, parameter = 0.5988)). The model of rate variation allowed some sites to be evolutionarily invariable ([+I], 12.0845% sites). The tree is drawn to scale, with branch lengths measured in relative number of substitutions per site. The analysis included 84 amino acid sequences, and the final data set had a total of 2106 positions.
(TIF)
The timetree shown was generated using RelTime. The estimated log likelihood value of the topology shown is -79253.3574. A discrete gamma distribution was used to model the differences in evolutionary rate amongst sites (five categories (+G, parameter = 0.6942)). The model of rate variation allowed some sites to be evolutionarily invariable ([+I], 13.5059% sites). The tree is drawn to scale, with branch lengths measured in relative number of substitutions per site. The analysis included 139 amino acid sequences, and the final data set had a total of 2299 positions.
(TIF)
(NEX)
Acknowledgments
The authors gratefully acknowledge the financial support from the Czech Science Foundation (grants no. 15–16050S and 17-23183S). The access to the computing and storage facilities owned by parties and projects contributing to the National Grid Infrastructure MetaCentrum provided under the programme “Projects of Large Infrastructure for Research, Development, and Innovations” (LM2010005) was highly appreciated, as was the access to the CERIT-SC computing and storage facilities provided under the programme “Center CERIT Scientific Cloud”, part of the Operational Program Research and Development for Innovations, reg. no. CZ.1.05/3.2.00/08.0144.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by the Czech Science Foundation (https://gacr.cz/en/), grants no. 15–16050S and 17-23183S, to DH.
References
- 1.Wellman CH. 2004. Origin, function and development of the spore wall in early land plants In: Hemsley AR, Poole I, eds. Evolution of plant physiology. Kew: Royal Botanic Gardens, 43–63. doi: 10.1104/pp.103.033068 [Google Scholar]
- 2.Wallace S, Fleming A, Wellman CH, Beerling DJ. 2011. Evolutionary development of the plant spore and pollen wall. AoB Plants 2011 plr027 doi: 10.1093/aobpla/plr027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Firon N, Nepi M, Pacini E. 2012. Water status associated processes mark critical stages in pollen development and functioning. Annals of Botany 109/7: 1201–1213. doi: 10.1093/aob/mcs070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quilichini TD, Grienenberger E, Douglas CJ (2015) The biosynthesis, composition and assembly of the outer pollen wall: A tough case to crack. Phytochemistry 113:170–182. doi: 10.1016/j.phytochem.2014.05.002 [DOI] [PubMed] [Google Scholar]
- 5.Williams JS (2008) Novelties of the flowering plant pollen tube underlie diversification of a key life history stage. Proc Natl Acad Sci U S A 105 (32):11259–11263 doi: 10.1073/pnas.0800036105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong XY, Hong ZL, Sivaramakrishnan M, Mahfouz M, Verma DPS. 2005. Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis. Plant Journal 42: 315–328. doi: 10.1111/j.1365-313X.2005.02379.x [DOI] [PubMed] [Google Scholar]
- 7.Nishikawa SI, Zinkl GM, Swanson RJ, Maruyama D, Preuss D. 2005. Callose (beta-1,3 glucan) is essential for Arabidopsis pollen wall patterning, but not tube growth. BMC Plant Biology 5: 9 doi: 10.1186/1471-2229-5-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paxson-Sowders DM, Dodrill CH, Owen HA, Makaroff CA (2001) DEX1, a novel plant protein, is required for exine pattern formation during pollen development in Arabidopsis. Plant Physiol 127: 1739–1749 [PMC free article] [PubMed] [Google Scholar]
- 9.Ariizumi T, Hatakeyama K, Hinata K, Inatsugi R, Nishida I, Sato S, Kato T, Tabata S, Toriyama K (2004) Disruption of the novel plant protein NEF1 affects lipid accumulation in the plastids of the tapetum and exine formation of pollen, resulting in male sterility in Arabidopsis thaliana. Plant J 39: 170–181 doi: 10.1111/j.1365-313X.2004.02118.x [DOI] [PubMed] [Google Scholar]
- 10.Guan YF, Huang XY, Zhu J, Gao JF, Zhang HX, Yang ZN (2008) RUPTURED POLLEN GRAIN1, a member of the MtN3/saliva gene family, is crucial for exine pattern formation and cell integrity of microspores in Arabidopsis. Plant Physiol 147: 852–863 doi: 10.1104/pp.108.118026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibalová A, Reňák D, Matczuk K, Dupl'áková N, Cháb D, Twell D, Honys D (2009) AtbZIP34 is required for Arabidopsis pollen wall patterning and the control of several metabolic pathways in developing pollen. Plant Molecular Biology 70 (5):581–601. doi: 10.1007/s11103-009-9493-y [DOI] [PubMed] [Google Scholar]
- 12.Backues SK, Korasick DA, Heese A, Bednarek SY (2010) The Arabidopsis Dynamin-Related Protein2 Family Is Essential for Gametophyte Development. Plant Cell 22 (10):3218–3231. doi: 10.1105/tpc.110.077727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reyes F, Leon G, Donoso M, Brandizzi F, Weber APM, Orellana A (2010) The nucleotide sugar transporters AtUTr1 and AtUTr3 are required for the incorporation of UDP-glucose into the endoplasmic reticulum, are essential for pollen development and are needed for embryo sac progress in Arabidopsis thaliana. Plant Journal 61 (3):423–435. doi: 10.1111/j.1365-313X.2009.04066.x [DOI] [PubMed] [Google Scholar]
- 14.Park J-I, Ishimizu T, Suwabe K, Sudo K, Masuko H, Hakozaki H, Nou I-S, Suzuki G, Watanabe M (2010) UDP-Glucose Pyrophosphorylase is Rate Limiting in Vegetative and Reproductive Phases in Arabidopsis thaliana. Plant and Cell Physiology 51 (6):981–996. doi: 10.1093/pcp/pcq057 [DOI] [PubMed] [Google Scholar]
- 15.Quilichini TD, Friedmann MC, Samuels AL, Douglas CJ (2010) ATP-binding cassette transporter G26 is required for male fertility and pollen exine formation in Arabidopsis. Plant Physiol 154: 678–690 doi: 10.1104/pp.110.161968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dobritsa AA, Lei Z, Nishikawa S, Urbanczyk-Wochniak E, Huhman DV, Preuss D, Sumner LW (2010) LAP5 and LAP6 encode anther-specific proteins with similarity to chalcone synthase essential for pollen exine development in Arabidopsis. Plant Physiol 153 (3):937–955. doi: 10.1104/pp.110.157446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J, Tian L, Sun M-X, Huang X-Y, Zhu J, Guan Y-F, Jia Q-S, Yang Z-N (2013) AUXIN RESPONSE FACTOR17 Is Essential for Pollen Wall Pattern Formation in Arabidopsis. Plant Physiology 162 (2):720–731. doi: 10.1104/pp.113.214940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibalová A, Steinbachová L, Hafidh S, Bláhová V, Gadiou Z, Michailidis C, Müller K, Pleskot R, Dupláková N, Honys D (2017) Characterization of pollen-expressed bZIP protein interactions and the role of ATbZIP18 in the male gametophyte. Plant Reprod 30 (1):1–17. doi: 10.1007/s00497-016-0295-5 [DOI] [PubMed] [Google Scholar]
- 19.Pacini E, Dolferus R. 2016. The trials and tribulations of the plant male gametophyte. Understanding reproductive stage stress tolerance. In Abiotic and Biotic Stress in Plants. Recent Advances and Future Perspectives; Shanker AK, Shanker C, Eds.; InTech: Rijeka, Croatia, 2016; pp. 703–754. [Google Scholar]
- 20.Ellinger D, Voigt CA. 2014. Callose biosynthesis in Arabidopsis with a focus on pathogen response: what we have learned within the last decade. Annals of Botany 114: 1349–1358. doi: 10.1093/aob/mcu120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X-Y, Kim J-Y. 2009. Callose synthesis in higher plants. Plant Signaling & Behavior 4: 489–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waterkeyn L. and Beinfait A. 1970. On a possible function of the callosic special wall in Ipomoea purpurea (L.) Roth. Grana 10: 13–20. [Google Scholar]
- 23.Falter C, Zwikiwics C, Eggert D, Blümke A, Naumann M, Wolff K et al. 2015. Glucanocellulosic ethanol: the undiscovered biofuel potential in energy crops and marine biomass. Scientific Reports 5: 13722, doi: 10.1038/srep13722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richmond TA, and Somerville CR. 2000. The cellulose synthase superfamily. Plant Physiology 124, 495–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verma DPS, Hong ZL. 2001. Plant callose synthase complexes. Plant Molecular Biology 47: 693–701. [DOI] [PubMed] [Google Scholar]
- 26.Hong Z, Delauney AJ, Verma DPS. 2001. A cell plate-specific callose synthase and its interaction with phragmoplastin. Plant Cell 13: 755–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU. 2005. A gene expression map of Arabidopsis thaliana development. Nat Genet 37 (5):501–506. doi: 10.1038/ng1543 [DOI] [PubMed] [Google Scholar]
- 28.Voigt CA, Schafer W, Salomon S. 2006. A comprehensive view on organ-specific callose synthesis in wheat (Triticum aestivum L.): glucan synthase-like gene expression, callose synthase activity, callose quantification and deposition. Plant Physiology and Biochemistry 44: 242–247. doi: 10.1016/j.plaphy.2006.05.001 [DOI] [PubMed] [Google Scholar]
- 29.Piršelová B, Matušíková I. 2013. Callose: The plant cell wall polysaccharide with multiple biological functions. Acta Physiologiae Plantarum 35: 635–644.·: doi: 10.1007/s11738-012-1103-y [Google Scholar]
- 30.Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. 2007. An “Electronic Fluorescent Pictograph” Browser for Exploring and Analyzing Large-Scale Biological Data Sets. PLoS ONE 2(8): e718 doi: 10.1371/journal.pone.0000718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie B, Deng YF, Kanaoka MM, Okada K, Hong ZL. 2012. Expression of Arabidopsis callose synthase 5 results in callose accumulation and cell wall permeability alteration. Plant Science 183: 1–8. doi: 10.1016/j.plantsci.2011.10.015 [DOI] [PubMed] [Google Scholar]
- 32.Stone BA, Clarke AE. 1992. Chemistry and physiology of higher plant1,3-b-glucans(Callose) In: Stone BA, Clarke AE (Eds). Chemistry and biology of (1,3)-b-glucans. Bundoora, Australia: La Trobe University Press, p. 365–429. [Google Scholar]
- 33.McCormick S. 2004. Control of male gametophyte development. PlantCell 16 (Suppl): S142–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Enns LC, Kanaoka MM, Torii KU, Comai L, Okada K, Cleland RE. 2005. Two callose synthases, GSL1 and GSL5, play an essential and redundant role in plant and pollen development and in fertility. Plant Molecular Biology 58: 333–349. doi: 10.1007/s11103-005-4526-7 [DOI] [PubMed] [Google Scholar]
- 35.Zhang C, Guinel FC and Moffatt BA. 2002. A comparative ultrastructural study of pollen development in Arabidopsis thaliana ecotype Columbia and male-sterile mutant Apt1-3. Protoplasma, 219, 59–71. [DOI] [PubMed] [Google Scholar]
- 36.Lu P, Chai M, Yang J, Ning G, Wang G, Ma H. 2014. The Arabidopsis CALLOSE DEFECTIVE MICROSPORE1 Gene Is Required for Male Fertility through Regulating Callose Metabolism during Microsporogenesis. Plant Physiology 164/4: 1893–1904. doi: 10.1104/pp.113.233387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cui W, Lee J-Y. 2016. Arabidopsis callose synthses CalS1/8 regulate plasmodesmatal permeability during stress. Nature Plants 16034. doi: 10.1038/NPLANTS.2016.34 [DOI] [PubMed] [Google Scholar]
- 38.Vatén A, Dettmer J, Wu S, Stierhof YD, Miyashima S, Yadav SR, et al. Callose biosynthesis regulates symplastic trafficking during root development. Dev Cell. 2011;21: 1144–1155. doi: 10.1016/j.devcel.2011.10.006 . [DOI] [PubMed] [Google Scholar]
- 39.Abercrombie JM, O'Meara BC, Moffatt AR, Williams JH. 2011. Developmental evolution of flowering plant pollen tube cell walls: callose synthase (CalS) gene expression patterns. Evolution and Development 2: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Töller A, Brownfield L, Neu C, Twell D, Schulze-Lefert P. 2008. Dual function of Arabidopsis glucan synthase-like genes GSL8 and GSL10 in male gametophyte development and plant growth. Plant Journal 54: 911–923. doi: 10.1111/j.1365-313X.2008.03462.x [DOI] [PubMed] [Google Scholar]
- 41.Maeda H, Song W, Sage T, DellaPenna D. 2014. Role of callose synthases in transfer cell wall development in tocopherol deficient Arabidopsis mutants. Frontiers in Plant Science 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang LJ, Chen XY, Rim Y, Han X, Cho WK, Kim SW, Kim JY. 2009. Arabidopsis glucan synthase-like 10 functions in male gametogenesis. Journal of Plant Physiology 166: 344–352. doi: 10.1016/j.jplph.2008.06.010 [DOI] [PubMed] [Google Scholar]
- 43.Thiele K, Wanner G, Kindzierski V, et al. 2009. The timely deposition of callose is essential for cytokinesis in Arabidopsis. Plant Journal 58: 13–26. doi: 10.1111/j.1365-313X.2008.03760.x [DOI] [PubMed] [Google Scholar]
- 44.Guseman JM, Lee JS, Bogenschutz NL, Peterson KM, Virata RE, Xie B, et al. 2010. Dysregulation of cell-to-cell connectivity and stomatal pattering by loss-of-fuction mutation in Arabidopsis chorus (glucan synthase-like8). Development 137: 1731–1741. doi: 10.1242/dev.049197 [DOI] [PubMed] [Google Scholar]
- 45.Xu T, Zhang C, Zhou Q, Yang ZN. 2016. Pollen wall pattern in Arabidopsis. Science Bulletin 61: 832–837. [Google Scholar]
- 46.Ellinger D, Voigt CA. 2014Callose biosynthesis in Arabidopsis with a focus on pathogen response: what we have learned within the last decade. Annals of Botany 114/6: 1349–1358. doi: 10.1093/aob/mcu120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang J, Tian L, Sun M-X, Huang X-Y, Zhu J, Guan Y-Y et al. 2013. Auxin Response Factor17 is essential for pollen wall pattern formation in Arabidopsis. Plant Physiology 162/2: 720–731. doi: 10.1104/pp.113.214940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schuette S, Wood AJ, Geisler M, Geisler-Lee J, Ligrone R, Renzaglia KS. 2009. Novel localization of callose in the spores of Physcomitrella patens and phylogenomics of the callose synthase gene family. Annals of Botany 103: 749–756. doi: 10.1093/aob/mcn268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Honys D, Twell D (2004) Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biology 5 (11). doi:R85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Majerská J, Schrumpfová Prochazková P, Dokládal L, Schořová S, Stejskal K, Obořil M, Honys D, Kozaková L, Polanská PS, Sýkorová E (2016) Tandem affinity purification of AtTERT reveals putative interaction partners of plant telomerase in vivo. Protoplasma. doi: 10.1007/s00709-016-1042-3 [DOI] [PubMed] [Google Scholar]
- 51.Fíla J, Záveská Drábková L, Gibalová A and Honys D. 2017. When simple meets complex; pollen and the -omics—Obermeyer G. and Feijó J.(ed.) Pollen Tip Growth—From Biophysical Aspects to System Biology. Springer International Publishing AG, pp. 247–292. [Google Scholar]
- 52.Craigon DJ, James N, Okyere J, Higgins J, Jotham J, May S (2004) NASCArrays: a repository for microarray data generated by NASC's transcriptomics service. Nucleic Acids Res 32 (Database issue):D575–577 doi: 10.1093/nar/gkh133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li C, Wong WH (2001) Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A 98 (1):31–36 doi: 10.1073/pnas.98.1.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li C, Wong WH (2001) Model-based analysis of oligonucleotide arrays: Model validation, design issues and standard error application. Genome Biol 2:R32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hafidh S, Breznenová K, Honys D (2012) De novo post-pollen mitosis II tobacco pollen tube transcriptome. Plant Signal Behav 7 (8):918–921. doi: 10.4161/psb.20745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hafidh S, Breznenová K, Ružička P, Fecikova J, Čapková V, Honys D (2012) Comprehensive analysis of tobacco pollen transcriptome unveils common pathways in polar cell expansion and underlying heterochronic shift during spermatogenesis. BMC Plant Biol 12:24 doi: 10.1186/1471-2229-12-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bokvaj P, Hafidh S, Honys D 2015. Transcriptome profiling of male gametophyte development in Nicotiana tabacum. Genomics Data 3: 106–111. doi: 10.1016/j.gdata.2014.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Honys D, Reňák D, Feciková J, Jedelský PL, Nebesářová J, Dobrev P, Čapková V (2009) Cytoskeleton-associated large RNP complexes in tobacco male gametopyte (EPPs) are associated with ribosomes and are involved in protein synthesis, processing and localisation. Journal of Proteome Research, 8(4): 2015–2031. doi: 10.1021/pr8009897 [DOI] [PubMed] [Google Scholar]
- 59.Jurečková JF, Sýkorová E, Hafidh S, Honys D, Fajkus J, Fojtová M (2017) Tissue-specific expression of telomerase reverse transcriptase gene variants in Nicotiana tabacum. Planta 245 (3):549–561. doi: 10.1007/s00425-016-2624-1 [DOI] [PubMed] [Google Scholar]
- 60.Sievers F. et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol., 7, 539 doi: 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Néron B, Ménager H, Maufrais C, Joly N, Maupetit J, Letort S et al. 2009. Mobyle: a new full web bioinformatics framework. Bioinformatics 25 (22): 3005–3011. doi: 10.1093/bioinformatics/btp493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Söding J (2005) Protein homology detection by HMM–HMM comparison. Bioinformatics 21: 951–960. doi: 10.1093/bioinformatics/bti125 [DOI] [PubMed] [Google Scholar]
- 63.Chen Y, Zou M, Cao Y. 2014. Transcriptome analysis of the Arabidopsis semi-in vivo pollen tube guidance system uncovers a distinct gene expression profile. J Plant Biol 57 (2):93–105. [Google Scholar]
- 64.Kumar S, Stecher G, and Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33:1870–1874. doi: 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Letunic I and Bork P. 2016. Interactive Tree Of Life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. doi: 10.1093/nar/gkw290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014 May 1; 30(9): 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tamura K, Battistuzzi FU, Billing-Ross P, Murillo O, Filipski A, Kumar S 2012. Estimating divergence time in large molecular phylogenies. PNAS 109/47: 19333–19338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bateman A., Birney E., Cerruti L., Durbin R., Etwiller L., Eddy SR. et al. 2002. The Pfam Protein Families Database Nucl. Acids Res. 30(1):276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Geer LY, Domarchev M, Lipman DJ, Bryant SH. 2002. CDART: protein homology by domain architecture. Genome Research 12/10: 1619–1623. doi: 10.1101/gr.278202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bailey TL, Bodén M, Buske FA, Frith M, Grant CE, Clementi L et al. 2009. MEME SUITE: tools for motif discovery and searching", Nucleic Acids Research, 37: W202–W208. doi: 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hu B, Jin J, Guo A-Y, Zhang H, Luo J. and Gao G. 2015. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics, 31(8):1296–1297. doi: 10.1093/bioinformatics/btu817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Borges F, Gomes G, Gardner R, Moreno N, McCormick S, Feijo JA, Becker JD (2008) Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiology 148 (2):1168–1181. doi: 10.1104/pp.108.125229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Honys D, Combe JP, Twell D, Čapková V (2000) The translationally repressed pollen-specific ntp303 mRNA is stored in non-polysomal mRNPs during pollen maturation. Sex Plant Reprod 13: 135–144. [Google Scholar]
- 74.Edwards, KD, Fernandez-Pozo N, Drake-Stowe K., M. Humphry M, Evans AD, Bombarely AD et al. 2017. A reference genome for Nicotiana tabacum enables map-based cloning of homeologous loci implicated in nitrogen utilization efficiency. BMC genomics 18: 448–462. doi: 10.1186/s12864-017-3791-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ward DM, Vaughn MB, Shiflett SL, White PL, Pollock AL, Hill J et al. 2005. The role of LIP5 and CHMP5 in multivesicular body formation and HIV-1 budding in mammalian cells. Biological Chemistry 280: 10548–10555. [DOI] [PubMed] [Google Scholar]
- 76.Douglas CM, Foor F, Marrinan JA, Morin N, Nielsen JB, Dahl AM et al. 1994. The Saccharomyces cerevisiae FKS1 (ETG1) gene encodes an integral membrane protein which is a subunit of 1,3-beta-D-glucan synthase. PNAS 91/26: 12907–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Williams JH, Taylor ML, O'Meara BC. 2014. Repeated evolution of tricellular (and bicellular) pollen. American Journal of Botany 101 (4):559–571. doi: 10.3732/ajb.1300423 [DOI] [PubMed] [Google Scholar]
- 78.Wickett N. J., Mirarab S., Nguyen N., Warnow T., Carpenter E., Matasci N., et al. 2014. Phylotranscriptomic analysis of the origin and early diversification of land plants. PNAS 111: E4859–E4868. doi: 10.1073/pnas.1323926111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gaudiner JC, Taylor NG, Turner SR 2003. Control of cellulose synthase complex localization in developing xylem. Plant Cell 15/8:1740–1748. doi: 10.1105/tpc.012815 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Highlighted domains are found in different plants.
(XLSX)
Sequence logos indicates main 50 conserved sequence patterns.
(DOCX)
The evolutionary history was inferred by using maximum likelihood; the tree with the highest log likelihood (-979085.0765) is shown. The analysis included a total of 4150 positions in the final data set. Main branches with circles are described in the text.
(TIF)
The timetree shown was generated using RelTime. The estimated log likelihood value of the topology shown is -57276.0114. A discrete gamma distribution was used to model the differences in evolutionary rate amongst sites (five categories (+G, parameter = 0.5505)). The model of rate variation allowed some sites to be evolutionarily invariable ([+I], 11.2557% sites). The tree is drawn to scale, with branch lengths measured in relative number of substitutions per site. The analysis included 105 amino acid sequences, and the final data set had a total of 2190 positions.
(TIF)
The timetree shown was generated using RelTime. The estimated log likelihood value of the topology shown is -91929.9006. A discrete gamma distribution was used to model the differences in evolutionary rate amongst sites (five categories (+G, parameter = 0.8543)). The model of rate variation allowed some sites to be evolutionarily invariable ([+I], 7.5727% sites). The tree is drawn to scale, with branch lengths measured in relative number of substitutions per site. The analysis included 127 amino acid sequences, and the final data set had a total of 2476 positions.
(TIF)
The timetree shown was generated using RelTime. The estimated log likelihood value of the topology shown is -54519.4579. A discrete gamma distribution was used to model the differences in evolutionary rate amongst sites five categories (+G, parameter = 0.5253)). The model of rate variation allowed some sites to be evolutionarily invariable ([+I], 0.7256% sites). The tree is drawn to scale, with branch lengths measured in relative number of substitutions per site. The analysis included 104 amino acid sequences, and the final data set had a total of 2274 positions.
(TIF)
The timetree shown was generated using RelTime. The estimated log likelihood value of the topology shown is -54506.2366. A discrete gamma distribution was used to model the differences in evolutionary rate amongst sites (five categories (+G, parameter = 0.5988)). The model of rate variation allowed some sites to be evolutionarily invariable ([+I], 12.0845% sites). The tree is drawn to scale, with branch lengths measured in relative number of substitutions per site. The analysis included 84 amino acid sequences, and the final data set had a total of 2106 positions.
(TIF)
The timetree shown was generated using RelTime. The estimated log likelihood value of the topology shown is -79253.3574. A discrete gamma distribution was used to model the differences in evolutionary rate amongst sites (five categories (+G, parameter = 0.6942)). The model of rate variation allowed some sites to be evolutionarily invariable ([+I], 13.5059% sites). The tree is drawn to scale, with branch lengths measured in relative number of substitutions per site. The analysis included 139 amino acid sequences, and the final data set had a total of 2299 positions.
(TIF)
(NEX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.