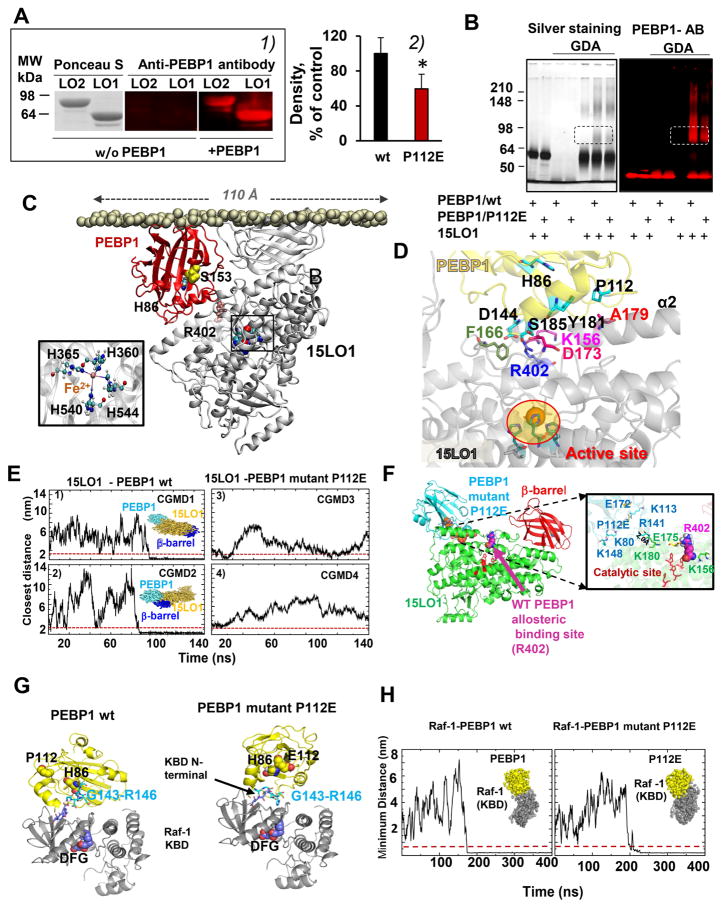
Figure 1. PEBP1 forms complexes with 15LO1 and 15LO2.
(A) 1) Far Western blotting gels showing binding of PEBP1 with 15LO. Ponceau S staining of proteins (left panel). Immuno-detection of PEBP1 bound to 15LO using anti-PEBP1 antibody (middle and right panels). 2) Quantitation of 15LO1 bound to wt PEBP1 and P112E PEBP1 with anti-15LO1 antibody (means±SD, *p < 0.05 vs. wt PEBP1, N=7/group.
(B) SDS-PAGE gels illustrating interactions of 15LO1 with wt PEBP1 and P112E PEBP1 revealed by cross-linking using glutaric dialdehyde (GDA). Proteins were detected by silver-staining or by anti–PEBP1 antibodies.
C) Computational modeling of human PEBP1/15LO1 complex. 15LO1 (gray) and PEBP1 (red) bound to the lipid bilayer are displayed, with the PL head phosphorus atoms in tan spheres, and 15LO1 R402, crucial for PEBP1/15LO1 interactions, in pink spheres. Inset: Coordination of Fe2+ at 15LO1 catalytic byH360, H365, H540 and H544.
(D) Close-up view of PEBP1/15LO1 interface. The interface between PEBP1 (yellow) and 15LO1 (gray) complex closely neighbors the AA binding site located near the catalytic site (highlighted in ellipse). K156, F166, D173, R402 in 15LO1, and P112, D144 and Y181 in PEBP1 make interfacial contacts that stabilize the PEBP1/15LO1 complex.
(E) Coarse-grained molecular dynamics (CGMD) simulations confirm the inability of the P112E to bind 15LO1. Four independent runs are displayed, two for wt PEBP1 (CGMD1 and 2), and two for the mutant (CGMD3 and 4). The ordinates denote the closest distance between the two proteins, originally separated by 20 Å. In CGMD1 and 2, stable bound poses are reached ~80–100 ns; the minimal distance attained and maintained during the remaining 150 ns. For the mutant, there is either no formation of complex (3), or transient binding (4) indicating weaker (if any) binding of P112E to 15LO1.
(F) Binding of P112E to 15LO1 in silico shows the weaker affinity and distinctive binding. Left: the optimal binding pose for mutant P112E is shifted away from the putative allosteric binding site near R402. Right: interfacial interactions. Wt PEBP1 exhibits several close contacts (see also Figure S5), which are broken due to the repulsion between mutated P112 (E112) and E175 on 15LO1. P112E and 15LO1 residues are colored as blue and green, respectively. E112 on PEBP1 and E175 on 15LO1 undergo side chain rotations in the mutant complex that weaken the intermolecular interactions.
(G) Binding of wt PEBP1 and P112E to Raf-1 kinase whose binding domain (KBD) (PDB id: 3c4c) (Tsai et al., 2008) was docked against human PEBP1 (left) and P112E mutant (right). Top docking poses of PEBP1 were in the vicinity of KBD N-terminus. Both wt (left) and P112E (right) utilize the region G143-R146 on PEBP1 for interacting with Raf-1 KBD (Deiss et al., 2012).
(H) CGMD of Raf-1 binding to wt PEBP1 (left) and P112E (right). In both cases, the complex was formed (distance less than 0.5 nm, indicated by dashed redline) at approximately 175ns (left) and 200ns (right).