Figure 3.
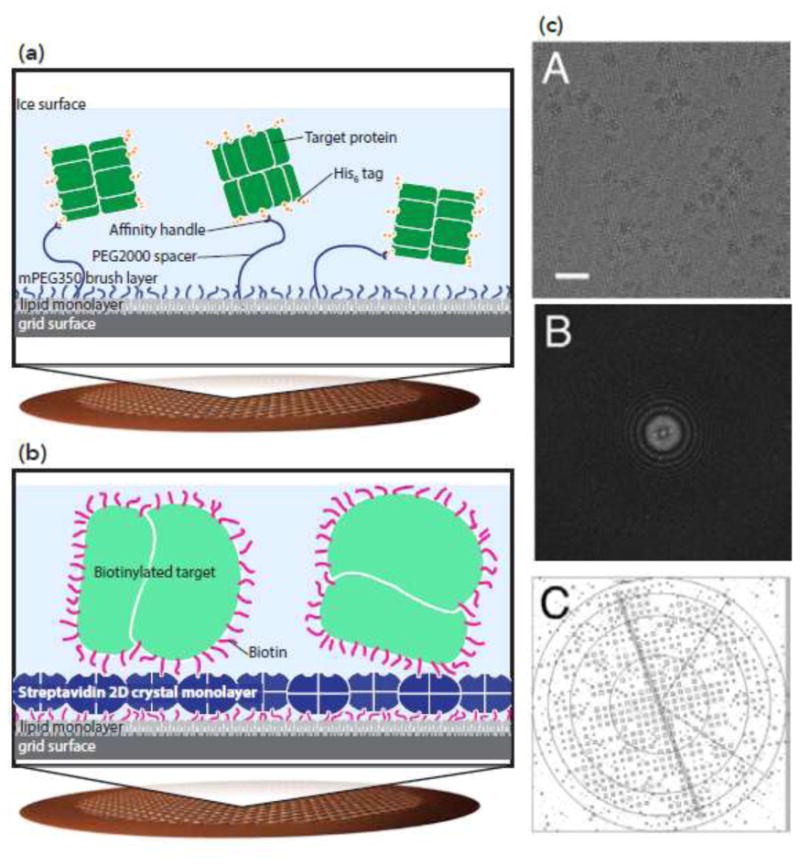
Affinity grids can control protein distribution and orientation. (a) Ni-NTA-based affinity grids. A copper mesh grid with holey carbon is functionalized with polyethylene glycol; long polymers with Ni-NTA are mixed with short polymers. Long polymers capture His-tagged proteins. Adapted from [65]. (b-c) Streptavidin-based affinity grids. (b) 2D crystals of streptavidin are layered on top of holey carbon grids. Biotinylated proteins stick to the streptavidin surface of the grid. (c) The streptavidin crystal can be seen on the surface of the grid, and density for the streptavidin crystal can be computationally removed. Top: cryo-EM micrograph of biotinylated ribosome on streptavidin affinity grid; center: Fourier transform of the image; bottom: IQ plot showing the diffraction pattern from the streptavidin crystal. From [67].
