Fig. 2.
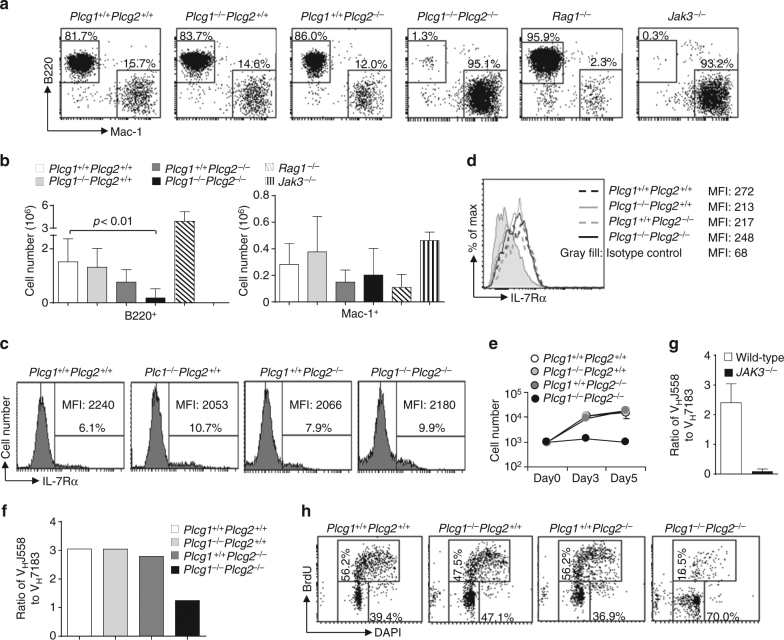
PLCγ1/PLCγ2 double deficiency impairs IL-7-mediated B cell progenitor functions. BM from Plcg1 +/+ Plcg2 +/+, Plcg1 −/− Plcg2 +/+, Plcg1 +/+ Plcg2 −/−, or Plcg1 −/− Plcg2 −/− mice were transplanted into lethally irradiated congenic wild-type CD45.1+ mice. Six to eight weeks after transplantation, Lin− BM cells from the recipients, Rag1 −/− or Jak3 −/− mice were cultured on OP9 cells plus IL-7. a, b IL-7-dependent B cell production. The cells were stained with anti-CD45.2, anti-B220, and anti-Mac-1 after 9 days of co-culture. Numbers indicate percentages of B220+ and Mac-1+ cells in the gated CD45.2+ population (a). Bar graphs show the numbers of CD45.2+B220+ and CD45.2+Mac-1+ cells (b). c, d IL-7R expression. BM cells from the recipients were stained with anti-IL-7Rα. Numbers indicate percentages of IL-7Rα+ cells in the gated CD45.2+Lin−population (c). After 9 days of co-culture plus Flt-3L, the cells from the recipients were stained with anti-CD45.2, anti-B220, and anti-IL-7Rα. Levels of IL-7Rα expression in the gated CD45.2+B220+ population are shown (d). e Expansion in response to IL-7. Fractions B and C of B cell progenitors (YFP+CD45.2+B220+CD43+CD24+) were sorted from the recipients and cultured with OP9 plus IL-7. On the indicated days after co-culture, the numbers of live cells were determined by trypan blue exclusion. f, g IL-7-mediated rearrangement of the distal VHJ558 family gene segments of IgH chain genes. YFP+CD45.2+B220+CD43+ or B220+IgM−CD43+ pro-B cells were sorted from the recipients (f) or from wild-type or Jak3−/− mice (g), respectively. The frequency of VHJ558 and VH7183 family usage in these progenitors was determined by high-throughput sequencing of the rearranged IgH genes. Bar graphs show the ratios of percentages of rearranged VHJ558 genes to those of rearranged VH7183 genes in the pro-B cells. h IL-7-mediated B cell progenitor proliferation. YFP+CD45.2+B220+CD43+CD24+ B cell progenitors (fractions B and C) were sorted from the recipients, and co-cultured with OP9 cells plus IL-7 for 3 days. BrdU incorporation and DAPI staining were used to determine proliferation. Numbers indicate percentages of BrdU+ cells in the gated live cells. Error bars show ± SEM. Data shown are obtained from or representative of 6 (a, b), 4 (c, h), or 2 (d–f) independent experiments or from three wild-type and 2 Jak3 −/− mice (g)