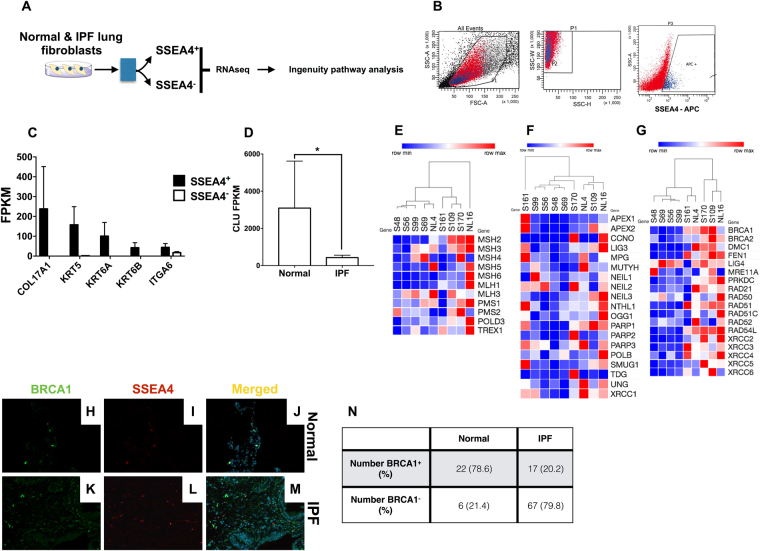
Figure 5.
Loss of transcripts for Clusterin, MMR, BER & DSB DNA repair pathway components in IPF SSEA4+ basal-like epithelial cells. (A) SSEA4+ and SSEA4− cells were sorted from normal and IPF stromal cultures, RNA was extracted and subjected to RNAseq analysis. Fold changes in IPF relative to normal were calculated and the results were uploaded onto ingenuity IPA for pathway analysis (A). (B) Shown is an representative sort plot depicting the forward and side scatter localization of SSEA4+ cells. (C) Normalized RNA sequencing FPKM values were mined for transcripts that were enriched in SSEA4+ relative to SSEA4− cells. Shown are enriched basal cell lineage markers in normal lung and IPF SSEA4+ cells. (D) Shown is the average CLU transcript FPKM values normal and IPF SSEA4+ cells. (E–G) Transcripts encoding for components of MMR (E), BER (F) and DSB (G) pathways were mined from SSEA4+ RNAseq data, clustered using Euclidean distance with complete linkage and heat maps were generated using Morpheus (Broad Institute). (H–M) Normal and IPF lung biopsies were stained with anti-SSEA4 and BRCA1 antibodies followed by fluorescent microscopic analysis. Representative images were taken at 200x magnification from two normal and four IPF lung biopsies stained with BRCA1 (H,K respectively), SSEA4 (I,L respectively) and the merged composites (J,M respectively). (N) The number of SSEA4+ cells showing both SSEA4 and BRCA1 staining was quantified, averaged, and the percentage of cells was calculated from 28 normal and 84 IPF SSEA4+ cells from 3 normal and 4 IPF lungs, respectively.