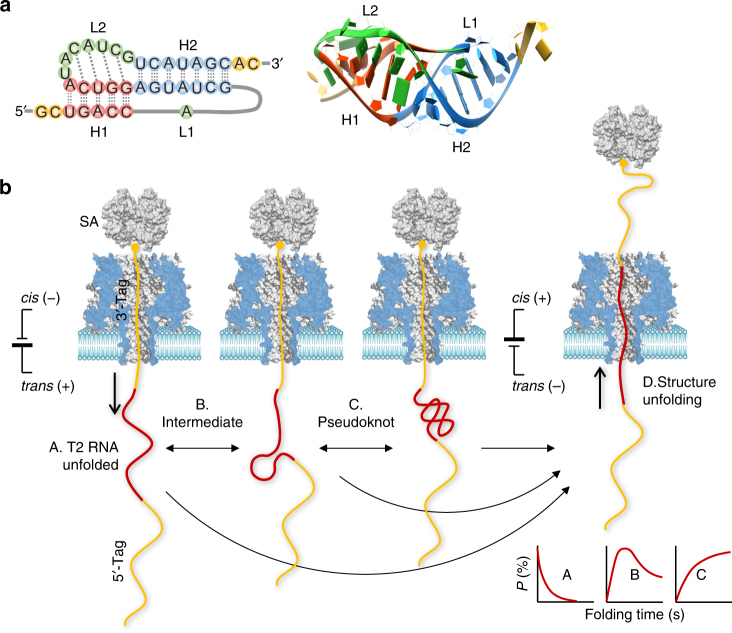
Fig. 1.
Single-molecule detection of RNA folding in a protein nanopore. a 2D (left) and 3D (right) structures of the pseudoknot from the 5ʹ end gene 32 messenger RNA of bacteriophage T2 (PDB id: 2tpk), which contains two helices, H1 (red) and H2 (blue), and two loops (green), L1 and L2. The base triple interactions between loops and helices (identified by RNAview70) and the coaxial stacking between the two helices are illustrated. b Nanopore detection of T2 RNA folding states and the folding pathway. The detection was facilitated by a complex probe containing a chimera of the 5ʹ-DNA tag, T2 RNA, and 3ʹ-DNA tag. The 3ʹ end is attached to streptavidin (SA) to immobilize the probe. Upon immobilization, the T2 RNA starts to fold from the single-stranded form in the trans solution. After a pre-defined folding time, the folding process is terminated at an intermediate state (B) or the pseudoknot structure (C) by using a negative voltage that disrupts the folding structure (D). The unfolding signatures reveal the folding state (B and C) and unfolding state (A). The folding time-dependent population of the identified folding states allows establishment of the pseudoknot folding pathway