Abstract
Mutations in the gene for dysferlin cause a degenerative disorder of skeletal muscle known as limb girdle muscular dystrophy 2B. To achieve gene delivery of plasmids encoding dysferlin to hind limb muscles of dysferlin knockout mice, we used a vascular injection method that perfused naked plasmid DNA into all major muscle groups of the hind limb. We monitored delivery by luciferase live imaging and western blot, confirming strong dysferlin expression that persisted over the 3-month time course of the experiment. Co-delivery of the follistatin gene, which may promote muscle growth, was monitored by ELISA. Immunohistochemistry documented the presence of dysferlin in muscle fibers in treated limbs, and PCR confirmed the presence of plasmid DNA. Because dysferlin is involved in repair of the sarcolemmal membrane, dysferlin loss leads to fragile sarcolemmal membranes that can be detected by permeability to Evan’s blue dye. We showed that after gene therapy with a plasmid encoding both dysferlin and follistatin, statistically significant reduction in Evan’s blue dye permeability was present in hamstring muscles. These results suggest that vascular delivery of plasmids carrying these therapeutic genes may lead to simple and effective approaches for improving the clinical condition of limb girdle muscular dystrophy 2B.
Keywords: dysferlin, muscle, naked DNA, follistatin, gene therapy, limb girdle muscular dystrophy 2B
Introduction
There are over 50 forms of hereditary muscular dystrophy due to mutations in different genes affecting muscle function.1 Most of these disease genes are recessive, making these disorders candidates for gene therapy by addition of a functional copy of the gene that is mutated.2 One of these recessive muscular dystrophies is limb girdle muscular dystrophy 2B (LGMD2B), caused by deficiency of dysferlin,3, 4 a protein involved in repair of the sarcolemmal membrane.5, 6 Mutations in the gene for dysferlin (DYSF) lead to a degenerative disorder of skeletal muscle, centered on muscles of the pelvic and shoulder girdles, with progressive involvement of distal limb muscles.3, 4, 5, 6, 7
Mouse models that lack dysferlin are available and have been used to demonstrate gene therapy for LGMD2B using a gene delivery strategy based on adeno-associated virus (AAV).6, 8, 9, 10, 11 Although promising results have been achieved in these studies, they also highlight some of the challenges of an AAV-mediated approach for LGMD2B. The dysferlin coding sequence is 6.2 kb long, but AAV is unable to package genes longer than 5 kb. Therefore, dual vector strategies8, 9, 10 or deleted forms of dysferlin11 have been used, which may reduce the efficiency or effectiveness of the therapy. In addition, an AAV gene therapy strategy presents problems with vector immunogenicity,12 vector loss, and the challenges and expense of large-scale viral vector production. The use of plasmid DNA as a vector could address these challenges because plasmid DNA has no size limit, is not immunogenic, is produced at a lower cost, and has the potential to integrate into the genome if an integration mechanism is provided.13
Jon Wolff and colleagues14 showed in 1990 that plasmid DNA provided surprisingly high levels of persistent gene expression when the DNA was introduced by intramuscular injection in mouse muscles. In order to deliver the DNA more broadly to multiple muscles, the Wolff group developed a method they called hydrodynamic limb vein injection.15 In this method, the hind limb circulation was transiently blocked by a tourniquet near the hip joint. Naked DNA was injected into the saphenous vein near the ankle joint in a volume of saline buffer sufficient to cause exit of the fluid and DNA from the blood vessels. This process of extravasation placed the DNA in direct contact with muscle fibers, where it was taken up and transcribed, leading to significant levels of delivered protein in the muscle fibers. The delivery process was simple, fast, and safe and lead to transfection of approximately 10% of muscle fibers in a single dose. Furthermore, the process could be repeated at intervals to build up incremental levels of therapeutic protein.16 The method was translated to monkey limbs with similar efficiency,17, 18 and the fluid delivery procedure was found to be safe and well tolerated in muscular dystrophy patients,19, 20 suggesting the possibility of clinical translation.
We experimented here with plasmid DNA-mediated vascular delivery in a mouse model of LGMD2B. Given the lack of size limit with plasmid vectors, it was possible to co-deliver the therapeutic dysferlin gene with other genes that might be beneficial. Therefore, we also demonstrated co-delivery of the 344-aa secreted isoform of the gene for follistatin (FST), which has a role in muscle anabolism.21 Follistatin has been shown to be beneficial in studies in mouse models of Duchenne muscular dystrophy, a severe form of muscular dystrophy, especially when co-delivered with the therapeutic dystrophin gene.22 Even though there is evidence that overproduction of follistatin alone may be harmful in LGMD2B mice,23 we hypothesized that muscle delivery by gene therapy of both dysferlin and follistatin on the same vector might be beneficial. In this study, we experiment with plasmid-mediated delivery of dysferlin or dysferlin plus follistatin in a mouse model of LGMD2B, assaying levels and distribution of gene expression by luciferase live imaging, western blotting, immunohistochemistry, and ELISA, presence of plasmid DNA by PCR, and benefit to muscle by the Evan’s blue dye (EBD) assay of muscle membrane permeability. These studies indicate that robust delivery of therapeutic proteins is possible by this method and may lead to beneficial changes in muscle physiology.
Results
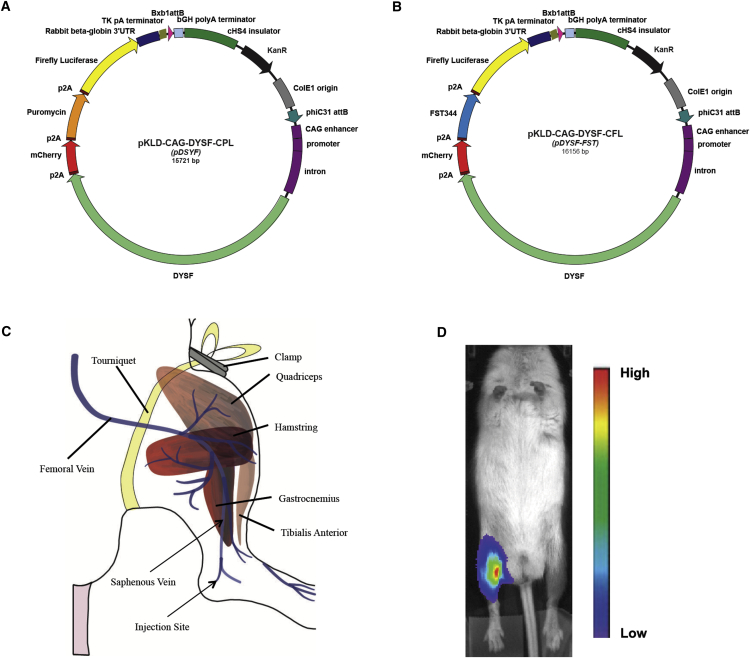
To carry out our plasmid gene therapy approach for LGMD2B, we utilized two therapeutic plasmids (Figures 1A and 1B). The plasmids carry the cDNA for human DYSF, a 6.2-kb coding sequence.24 To obtain strong expression of DYSF, we used the CAG promoter enhancer, encompassing the widely expressed CMV enhancer, chicken β-actin promoter, and a hybrid intron.25, 26 We also included the cHS4 chicken insulator element to combat silencing of gene expression.27 The rabbit β-globin 3′ UTR, thymidine kinase polyA sequences, and bovine growth hormone polyA were used for transcription termination. In addition, marker genes encoding mCherry, puromycin resistance, and firefly luciferase, linked by p2A sequences for coordinated expression, were part of the dysferlin expression cassette present in pKLD-CAG-DYSF-CPL, abbreviated pDYSF (Figure 1A).24, 28 To obtain a plasmid that co-expressed DYSF and FST, we removed the puromycin resistance gene from pDYSF, which was not needed in these experiments, and substituted DNA sequences encoding the 344-aa isoform of human follistatin, yielding pKLD-CAG-DYSF-CFL, abbreviated pDYSF-FST (Figure 1B).
Figure 1.
Plasmids and DNA Injection
(A) Map of plasmid pKLD-CAG-DYSF-CPL (pDYSF) expressing human dysferlin. (B) Map of plasmid pKLD-CAG-DYSF-CFL (pDYSF-FST) expressing human dysferlin and follistatin. (C) Schematic diagram illustrating blood vessels and muscles involved in vascular muscle perfusion method. (D) Luciferase live imaging photo of BLAJ/NRG mouse legs 4 weeks after injection with pDYSF (left) and HBSS (right), shown with a bioluminescence intensity gradient.
To achieve widespread delivery of therapeutic plasmids to hind limb muscles, we applied the hydrodynamic limb vein injection method described by Hagstrom et al.,15 which utilizes naked DNA and distributes it to all the major muscle groups of the mouse hind limb. As diagrammed in Figure 1C, we transiently blocked the hind limb circulation by clamping a rubber tourniquet as high on the leg as possible. 400 μg plasmid DNA in Hank’s balanced salt solution (HBSS) buffer were injected into the saphenous vein near the ankle using a needle attached to a syringe pump for rapid (8 mL/min) and controlled delivery of the fluid. Introducing the DNA in a volume of ∼1 mL to the leg in the presence of the tourniquet forced the fluid to exit the vasculature, where the plasmid DNA came in contact with muscle fibers, which are competent to take up naked DNA. Successful delivery was monitored by observing transient swelling of the limb as well as distribution of the blue dye that was included in the injection fluid. To detect expression of the plasmid DNA after injection, we followed expression of the firefly luciferase gene on the therapeutic plasmids by luciferase live imaging. As shown in Figure 1D, a bright luciferase signal was seen in legs injected with plasmid DNA, whereas no signal was seen in legs receiving buffer alone or elsewhere in the body.
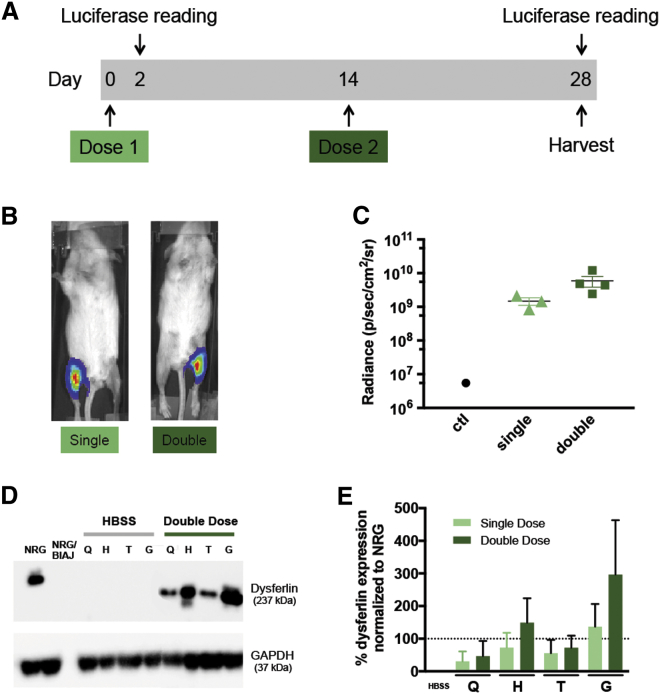
The low immunogenicity of naked plasmid DNA itself allows the potential for multiple doses to be administered. The Wolff group16 established that at least six doses of plasmid DNA could be given sequentially, with corresponding increases in the amount of protein delivered and the number of fibers transfected. In an initial gene therapy experiment, we tested the effect of two doses of DNA versus a single dose. As shown in Figure 2A, mice were given a single dose of pDYSF in one leg; then, 2 weeks later, a second dose of pDYSF was given to half of the animals in the same legs. The contralateral legs were injected with HBSS alone. All animals were harvested 4 weeks after the first injection. Luciferase bioluminescence live imaging was done on day 2 to verify injection quality, then again just before muscles were harvested. The mice were approximately 6 months old at the time of injection and approximately 7 months old at the end of the experiment. As shown in Figure 2B, strong luciferase signals were observed in legs that received DNA. Figure 2C indicates that the average luciferase signal tended to be stronger after 2 doses than after a single dose, as expected.
Figure 2.
Comparison of Single and Double Vascular Injections of pDSYF in BlAJ/NRG Mice in a 4-Week Experiment
(A) Timeline of the experiment; all mice received a first injection of plasmid pDYSF at day 0, whereas half of the mice also received a second injection of pDYSF at day 14. Luciferase live imaging was performed 2 and 28 days after injections. All mice were harvested at day 28. (B) Representative luciferase live images of mice 28 days after single (left) or double (right) doses of pDYSF. (C) Bioluminescence radiance values were graphed for control (HBSS alone) and single and double doses of pDYSF. Although double dose values trended higher, the difference in levels observed between the single and double dose groups was not statistically significant. (D) Western blot images of protein extracted from the four hind limb muscle groups from two representative legs after injection with either HBSS or a double dose of pDYSF. NRG represents the wild-type level of dysferlin, whereas NRG/BlAJ represents the null level of dysferlin in BlAJ/NRG mice. (E) Western blot band quantification of all the data showed an upward trend in dysferfin expression in the double dose groups that was not statistically significant. The dotted line represents 100% of wild-type levels of dysferlin. Q, quadriceps; H, hamstring; T, tibialis anterior; G, gastrocnemius. Data are mean ± SEM, with n = 3 to 4.
To quantify the levels of dysferlin protein present in the treated muscles, western blots were performed on protein extracted from the quadriceps, hamstring, tibialis anterior, and gastrocnemius muscles, and dysferlin levels were visualized by western immunoblotting with an antibody that recognized the 237-kDa dysferlin protein (Figure 2D). Equal amounts of protein (25 μg) were loaded in each lane, and the 37-kDa GAPDH protein was imaged as a loading control. Protein extracted from the parental NRG mice, which were wild-type for dysferlin, was used as a positive control representing the normal wild-type level of dysferlin. Protein extracted from untreated BlAJ/NRG mice was used as a negative control and contained no detectable dysferlin, in keeping with the null phenotype of the mouse model.8 Amounts of dysferlin protein present in legs treated with buffer or pDYSF were quantified by normalizing to GAPDH and using the NRG sample to represent 100%. Average amounts of dysferlin present as a percentage of wild-type levels are shown in Figure 2E for the legs receiving a single or double dose of plasmid DNA. We generally observed more dysferlin protein in the gastrocnemius and hamstring muscles than in the tibialis anterior and quadriceps muscles, and there was a trend toward higher levels of dysferlin protein after two doses. However, because even a single dose yielded values averaging 25 to >100% of normal, we decided to administer just a single dose of plasmid DNA in further experiments.
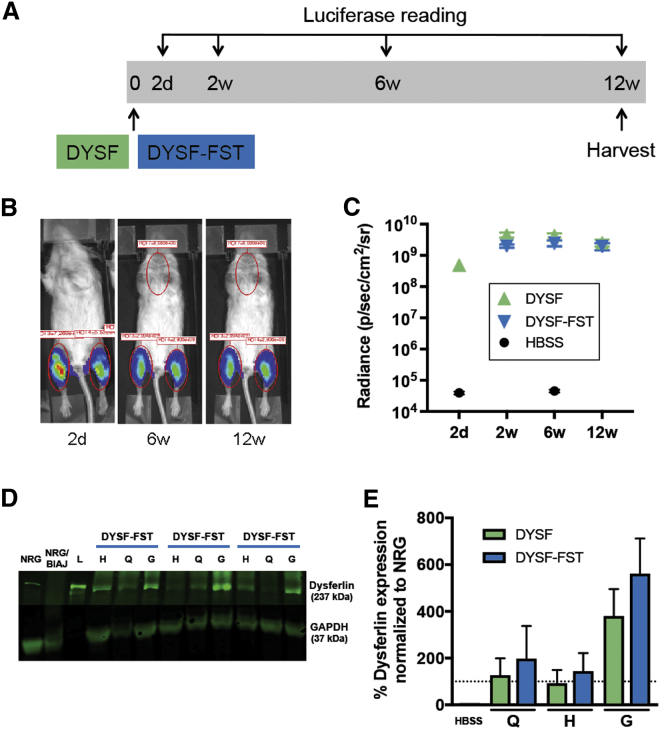
To test the durability of gene expression over a longer time period and to measure possible benefit of the treatment, we carried out a DYSF gene therapy experiment that lasted approximately 3 months. In addition to mice that received the pDYSF plasmid, we included a group of mice that were treated with a plasmid, pDYSF-FST, carrying both the DYSF gene and the gene encoding human FST to test the possible benefit of that combination. As shown in Figure 3A, the experimental scheme involved a single dose of plasmid DNA or HBSS buffer in each leg, with luciferase live imaging done 2 days after treatment to monitor injection success and at 2, 6, and 12 weeks to examine retention of gene expression. Two mice were treated with HBSS buffer alone, 4 with pDYSF, and 6 with pDYSF-FST. Each group of mice contained both genders, and the mice were aged 8–9 months at the start of the experiment and 11–12 months at the end. We deliberately used older animals to simulate patients in whom the disease process was well underway.7, 29
Figure 3.
Vascular Injection of pDYSF or pDYSF-FST in BlAJ/NRG Mice in a 12-Week Experiment
(A) Timeline of the experiment; all mouse legs received a single vascular injection of either HBSS alone, pDYSF, or pDYSF-FST at day 0, and hind limb muscles were collected at 12 weeks. Luciferase live imaging was performed at 2 days, 6 weeks, and 12 weeks after injection. (B) Representative luciferase images of the same mouse at 2 days, 6 weeks, and 12 weeks after injection with pDYSF-FST. (C) Similar levels of bioluminescence were observed in mice injected with pDSYF and pDYSF-FST. These levels were persistent over the time course of the experiment. (D) Representative western blot images of three mouse legs injected with pDYSF-FST; the H, Q, and G muscle groups were analyzed. NRG represents the wild-type level of dysferlin, whereas NRG/BlAJ represents the null level of dysferlin in BlAJ/NRG mice. Lane L is the protein size ladder. (E) Western blot band quantification showed no significant difference in dysferfin expression between the two plasmids FST. The dotted line represents 100% of wild-type levels of dysferlin present in NRG mice. Q, quadriceps; H, hamstring; G, gastrocnemius. Data are mean ± SEM, with n = 4–13.
Figure 3B shows images of a representative mouse treated with pDYSF-FST 2 days, 6 weeks, and 12 weeks after DNA injection. In Figure 3C, we graphed all the luciferase data, revealing that pDYSF and pDYSF-FST plasmids gave rise to similar levels of luciferase expression. Luciferase expression rose between 2 days and 2 weeks and then was stable for the 12-week duration of the experiment. The levels of dysferlin protein present in the quadriceps, hamstring, and gastrocnemius muscles were evaluated by western blot. Figure 3D shows a representative western blot for three legs that received pDYSF-FST. Dysferlin values were normalized using GAPDH and plotted as a percentage of normal levels of dysferlin in Figure 3E. Both pDYSF and pDYSF-FST expressed similar levels of dysferlin that averaged ∼100% or more of normal levels, with the highest levels in the gastrocnemius muscles. The delivery efficiency for the different muscle groups followed a similar order, as we observed in the previous experiment, although the absolute amounts of dysferlin protein trended higher in the longer experiment (Figure 2E compared with Figure 3E).
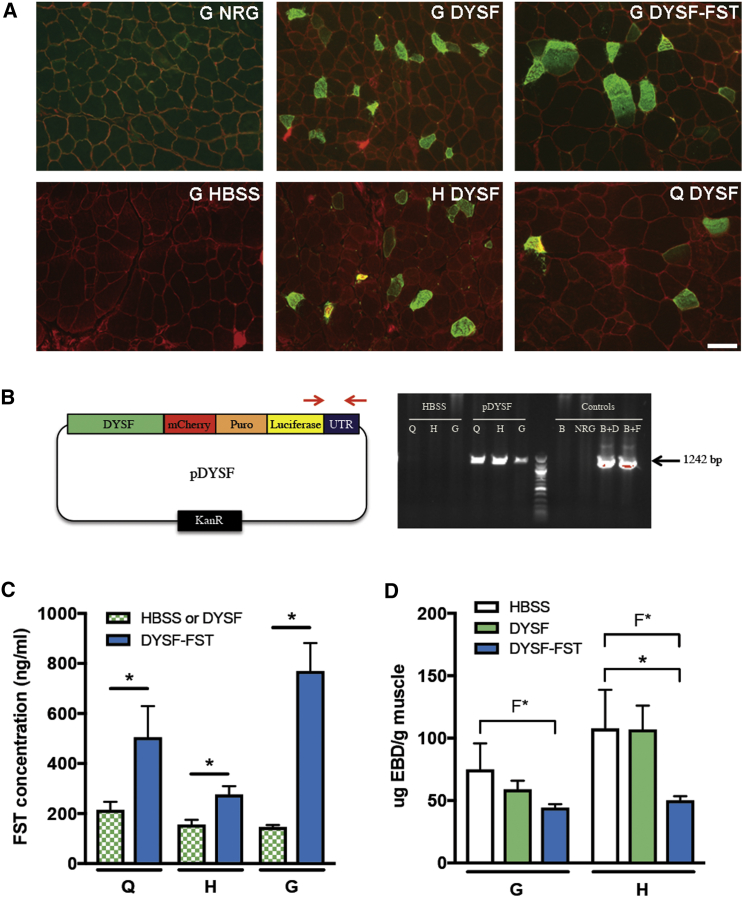
The muscles in this experiment were also analyzed by immunohistochemistry (Figure 4A). All sections were stained with laminin to demarcate the positions of muscle fibers. In the NRG-positive control, dysferlin was visible near the sarcolemmal membrane. In the BlAJ/NRG mouse model of LGMD2B, no dysferlin staining was visible, consistent with the null phenotype of these animals,8, 29 and no dysferlin staining was visible in muscles that received only HBSS. Sections from gastrocnemius, hamstring, and quadriceps muscles from legs that received pDYSF showed dysferlin-positive fibers, generally brighter than normal levels of dysferlin, with the presence of cytoplasmic vesicles, as has also been commonly observed in an AAV-mediated gene therapy experiment in mice.8, 9, 10, 11 A gastrocnemius muscle that received pDYSF-FST is also shown. These sections represent some of the highest densities of dysferlin-positive fibers we observed, with fiber counts revealing that 10%–12% of fibers were positive for a single dose. Because not all sections had this level of dysferlin-positive fibers, the overall density of positive fibers for this single dose was <10%.
Figure 4.
Further Analysis of 12-Week Gene Therapy Experiment in BlAJ/NRG Mice
(A) Immunohistochemistry staining on G, H, and Q muscle sections harvested 12 weeks after vascular injection of HBSS alone, pDYSF, or pDYSF-FST. Laminin is stained red, and dysferlin is stained green. A section from the positive control NRG mouse is also shown. Bar, 50 μm. (B) Presence of plasmid DNA was detected by PCR in muscles injected with HBSS alone, pDYSF, or pDYSF-FST. Left diagram shows the position of the primers used to detect an identical 1,242-bp region of DNA present on pDYSF and pDYSF-FST. Representative PCR lanes show DNA extracted from Q, H, and G muscles from legs treated with either HBSS alone or pDYSF-FST. Control lanes are B = BlAJ/NRG, NRG; B+D = BlAJ/NRG + pDYSF; and B+F = BlAJ/NRG + pDYSF-FST. (C) Follistatin concentration was quantified by ELISA from muscles injected with HBSS, pDYSF, or pDYSF-FST. Legs injected with pDYSF-FST expressed significantly more follistatin than legs injected with HBSS alone or pDYSF (pooled for statistical purposes) 28 days after the injections. (D) Concentration of Evan’s blue dye was significantly lower in hamstring muscles of legs injected with pDYSF-FST compared to legs injected with HBSS alone or pDYSF 12 weeks after injections. Variability was also significantly lower in muscles injected with pDYSF-FST compared to HBSS alone or pDYSF-injected muscles, as determined by F test. Q, quadriceps; H, hamstring; G, gastrocnemius. Data are mean ± SEM, with n = 4–8 and *p < 0.05.
A separate experiment was performed, in which ∼7-month-old mice received one dose of HBSS buffer, pDYSF, or pDYSF-FST. Muscles were harvested after 1 month to analyze the presence of plasmid DNA by PCR and the levels of follistatin protein present in the muscles by ELISA. We performed PCR on DNA extracted from these muscle samples using primers that detected luciferase plasmid sequences not present in the genome. As shown in Figure 4B, a strong PCR band was detected in legs that received plasmid DNA, but not in those that received only HBSS buffer, verifying the existence of plasmid DNA in these muscles. As shown in Figure 4C, levels of follistatin were significantly higher in muscles that received pDYSF-FST compared to those receiving pDYSF or HBSS, which reflected only the basal level of endogenous follistatin present in mouse muscles.
To analyze whether we could detect any improvement in the condition of the treated muscles, we injected Evan’s blue dye into the mice intraperitoneally 1 day before harvest of the muscles in the 12-week experiment. Evan’s blue dye is able to enter muscle fibers only if their sarcolemma is permeable, a condition increasingly present with progression of LGMD2B due to the membrane repair defect present in dysferlin-deficient muscle fibers. We showed that the hamstring muscle was the most permeable hind limb muscle to Evan’s blue dye at age matching of these mice.29 When the gastrocnemius and hamstring muscles were analyzed for levels of Evan’s blue dye, we found a trend downward in the treated muscles, with a statistically significant drop in Evan’s blue dye uptake present in hamstring muscles treated with pDYSF-FST (Figure 4D). Furthermore, the animal-to-animal variability, as determined by F test, was also lower in muscles that received pDYSF-FST, and this difference was statistically significant (Figure 4D).
Discussion
In this study, we demonstrated a gene therapy using naked plasmid DNA that distributed substantial amounts of therapeutic protein to hind limb muscles in a mouse model of LGMD2B. By performing a relatively simple procedure of vascular injection of DNA in the presence of a tourniquet to block circulation transiently, amounts of therapeutic dysferlin protein approaching or exceeding normal levels were detected in hind limb muscles by western blot 3 months after treatment. Immunohistochemistry confirmed the presence of dysferlin in muscle fibers, and PCR confirmed the presence of plasmid DNA. Luciferase live imaging provided a convenient surrogate marker to evaluate the strength and durability of gene expression of the delivered genes.
Previously, this vascular delivery method had been applied to mdx mouse models of Duchenne muscular dystrophy by the Wolff group.16 Generally, similar results were seen in the levels of gene expression observed and the preferential delivery to similar muscle groups, including the gastrocnemius and hamstring. In those studies, plasmids expressing full-length dystrophin provided stable dystrophin expression for the lifetime of the mice.16 Protection from Evan’s blue dye permeability was seen when about 20% of normal levels of dystrophin were present, distributed among 11%–16% of fibers.16 Repeat delivery was used to increase the amount of dystrophin protein and percentage of dystrophin-expressing fibers. Repeated delivery was found to increase the therapeutic protein level in a linear fashion, whereas the number of fibers transfected rose at a slower rate,16 presumably due to repeat delivery to some fibers, perhaps due to their proximity to blood vessels. Our results show that this methodology can be extended to other forms of muscular dystrophy, with similar results.
This approach contrasts with the current focus on AAV-mediated delivery for muscle gene therapy. Because it is uncertain whether the AAV approach will be sufficient, developing alternative gene therapy strategies may offer valuable options for patients. The primary advantage of the AAV system is that it can provide for efficient delivery to tissues via a receptor-mediated process involving the viral capsid that has often worked well in mice. By the same token, the presence of a viral capsid of fixed dimensions and composed of foreign proteins also creates limitations. Plasmid DNA has no size limit and, as demonstrated here, can accommodate the full-length dysferlin coding sequence. By contrast, AAV is a small, packaged virus with a fixed size limit of ∼4.7 kb. The full-length dysferlin coding sequence is 6.2 kb long and will not fit in AAV vectors. As a result, partial dysferlin sequences or dual vector approaches that require recombination in recipient cells are being used in AAV gene therapy development for dysferlinopathy.8, 9, 10, 11 However, dual vector approaches compromise efficiency and increase complexity, whereas partial fragments of the protein may have only partial activity. Use of the full-length coding sequence is more desirable and feasible with the plasmid system.
Another potential advantage of the plasmid system is lesser immune response. The AAV capsid stimulates an immune response in humans, and most humans have pre-existing antibodies to AAV, which may complicate treatment by inactivation of incoming virus.12 Moreover, after an AAV-based gene therapy is administered, a neutralizing antibody response to the AAV capsid may interfere with further treatment. The need for re-treatment is likely for muscular dystrophy because the muscle target is large and AAV gene therapy may not be permanent. By contrast, it has been shown that plasmid can be re-administered multiple times.16
An additional potential advantage of a plasmid system is the ability to integrate the plasmid covalently into the genome, for example, by using phiC31 integrase or another integration system.13, 30, 31 Integrated plasmid DNA becomes a permanent part of the genome, which may prolong therapeutic gene expression. Lack of integration of most of the AAV vector population means that therapeutic gene expression could diminish over time. AAV also has little ability to correct muscle stem cells, which are dividing cells that rapidly lose unintegrated DNA.32 Ease of manufacture is another potential advantage of a plasmid system. It is relatively easy and cheap to make plasmid DNA from bacteria. By contrast, the viral manufacturing and purification process is time consuming and expensive. A potential disadvantage of plasmid delivery is less efficient delivery. However, it should be noted that although AAV delivery in mouse muscle is efficient, it may be less efficient in humans due to a larger size and more potent immune response. A conceivable potential strategy is to administer an initial dose of AAV vector to achieve widespread coverage, followed by vascular injection of plasmid to boost expression over time.
Our results suggest that a large amount of dysferlin protein was made in a subset of fibers. It remains possible that low levels of dysferlin expression in additional fibers were obscured by the strong expression in some fibers. This uneven distribution of therapeutic protein may not be ideal for the types of structural or enzymatic proteins involved in many forms of muscular dystrophy. On the other hand, this pattern of distribution is attractive for delivery of secreted proteins. Follistatin is a small, conserved protein that is secreted locally and binds to and inhibits myostatin, a transforming growth factor β (TGF-β) family member that is expressed almost exclusively in developing and mature muscle and acts to shut down muscle growth.21 Follistatin can stimulate muscle growth even in myostatin null mice by inhibiting additional TGF-β family members, including activin.33 The 344-aa isoform of human FST that we used here has been tested in a human clinical trial that claimed a possible therapeutic benefit in Becker muscular dystrophy patients.34 Favorable distribution of locally secreted follistatin by the vascular delivery method we employed may help explain why the plasmid that encoded both dysferlin and follistatin appeared to be more therapeutically effective than a plasmid with dysferlin alone.
Expression of dysferlin in 100% of fibers is not required for clinical efficacy. For example, in a study of delivery of a truncated dysferlin gene with AAV, systemic delivery led to expression of dysferlin in ∼10% of muscle fibers in the gluteal muscles, but this was sufficient for histological and functional recovery.11 Although plasmid delivery to muscle by this vascular method is restricted to the area delineated by the tourniquet and does not include the heart, the heart is rarely affected in LGMD2B1, 2 and is not a critical target in this disorder. Furthermore, cardiac avoidance is an asset in some forms of muscular dystrophy, like LGMD2A, in which expression of the therapeutic calpain3 protein is toxic in the heart.35
In follow-up studies, it would be advantageous to treat younger animals and wait a longer period of time before evaluation. Such a protocol would provide a more rigorous test of safety and might result in more dramatic results from the gene therapy, permitting documentation of clinical benefits through muscle function testing. Furthermore, immune-competent Bl/AJ mice will be used in order to help predict any immune response resulting from therapeutic plasmids, from which marker genes such as luciferase will be removed. It is also of interest to test delivery of follistatin alone to better evaluate the benefit we saw in this study of a plasmid carrying both dysferlin and follistatin. It would be informative to evaluate muscle mass in those studies because increase in muscle mass is a documented feature of follistatin activity.21, 22, 23 It may also be beneficial to employ genomic integration of the plasmid. Genomic integration of plasmid DNA with phiC31 integrase, using a different delivery method and experimental setting, was shown to increase levels and duration of therapeutic protein expression and improve its distribution in muscle fibers.13 If the phiC31 integrase system also works to bring about sequence-specific genomic integration in this system, it may offer a simple, attractive addition to this gene delivery approach.30, 31
Vascular delivery of naked DNA has been extended to larger species than mice, including pigs and non-human primates.17, 18, 36 In particular, 184 primate limbs were injected by the Wolff group, including upper and lower limbs; all animals recovered quickly, without obvious side effects or deficits to muscles, nerves, or blood vessels.17, 18 Furthermore, there was little attenuation in efficiency of delivery in the larger animals compared to the results in mice. This is important because gene therapy methods often fail to translate well during the thousand-fold scale-up from mouse to human. Furthermore, high volume injection of fluid in lower and upper limbs was tested in human clinical trials in muscular dystrophy patients and was found to be safe, feasible, and well tolerated.19, 20 This delivery method clearly provides long-term expression of the therapeutic gene because we carried out a 12-week experiment and observed abundant dysferlin expression, a protein with a half-life of only ∼4–6 hours.37 These results suggest that this gene therapy approach may have potential for clinical application and could also be extended to further forms of limb girdle muscular dystrophy, such as LGMD2A and LGMD2D.
Materials and Methods
Plasmids and DNA
The therapeutic plasmid carrying the dysferlin coding sequence pKLD-CAG-DYSF-CPL was constructed as described.24 Briefly, the plasmid contains the human dysferlin cDNA cloned into the pKLD-CAG backbone,28 along with a gene expression cassette encoding mCherry, puromycin resistance, and firefly luciferase connected by p2A skipping peptides. To construct a derivative that also encoded follistatin, the puromycin resistance gene in pKLD-CAG-DYSF-CPL was replaced with the coding sequence for the 344-aa isoform of human follistatin, synthesized as a gBlock gene fragment (Integrated DNA Technologies, Coralville, IA) using the AgeI restriction enzyme site. Plasmid DNA used for animal injections was purified with an endotoxin-free maxi- or giga-prep kit (Macherey-Nagel, Düren, Germany) following the manufacturer’s instructions.
Mice
We utilized dysferlin null A/J mice that were crossed onto the C57BL/6 background to make the Bl/AJ strain.6, 8 Bl/AJ mice (B6.A-Dysfprmd/GeneJ, stock number 012767) were provided by the Jackson Laboratory (Bar Harbor, ME) from a colony maintained by the Jain Foundation. To obtain immune-deficient mice, we crossed the Bl/AJ mice with NRG mice that were deficient in B, T, and NK cells.38 NRG mice (NOD.Cj-Rag1tm1Mom Il2rgtm1Wjl/Szj, stock number 007799) were purchased from Jackson Laboratory. The resulting BlAJ/NRG mice were characterized in detail29 and donated to Jackson Laboratory as stock number 029663 (NOD.Cg-Rag1tm1MomDysfprmdIl2rgtm1Wjl/McalJ). The Stanford Administrative Panel on Laboratory Animal Care approved all procedures performed on animals. The Stanford Comparative Medicine program is accredited by the Assessment of Laboratory Animal Care International.
Vascular Injection
Plasmid DNA was injected into mouse hind limbs using the vascular delivery procedure described by Hagstrom et al.15 Briefly, mice were anesthetized with a ketamine-xylazine cocktail (K: 10 mg/mL, X: 1 mg/mL), a tourniquet was placed at the proximal joint of the leg, and a small incision was made near the ankle joint. 400 μg plasmid DNA in 1 mL HBSS (Thermo Fisher Scientific, Waltham, MA) or 1 mL HBSS buffer alone were injected into the saphenous vein using a 10-mL Luer-Lok syringe (Becton Dickinson, Franklin Lakes, NJ) attached to a 33G needle (TSK Laboratories, Tochigi, Japan) at a rate of 8 mL/min using a Legato 210 syringe pump (KD Scientific, Holliston, MA). The needle was withdrawn, pressure was applied for 1 min, the tourniquet was released, and the incision was sutured. Patent blue dye (0.25 mg/mL; Thermo Fisher Scientific) was included in the injection fluid to monitor injection success.
Luciferase Live Imaging
Mice to be imaged were anesthetized with 1.5% isofluorane through a nose cone. Luciferin substrate (1 mg/mL PBS; Biosynth, Itasca, IL) was injected into the intraperitoneal cavity (100 μL/10 g body weight). 10 min after luciferin injection, luminescence was detected using an in vivo imaging system (IVIS Spectrum Pre-Clinical In Vivo Imaging System, PerkinElmer, Waltham, MA) and associated software for quantitation. Luminescence images were acquired at exposure times of 1 s or less. Luminescence was quantified within regions of interest in units of photons/s/cm2.
Western Blots
The quadriceps, hamstring, gastrocnemius, and tibialis anterior muscle groups were dissected from mouse hind limbs. Muscle lysates were prepared with RIPA buffer supplemented with HALT Protease Inhibitor Cocktail (Thermo Fisher Scientific) according to the manufacturer’s protocol. The supernatant-containing protein extract was denatured with Laemmli Sample Buffer (Bio-Rad, Hercules, CA) supplemented with 100 mM DTT. In each lane of a 10% TRIS-glycine SDS-PAGE gel (Bio-Rad), 25 μg protein extract was electrophoresed at 70 V in ice cold PAGE running buffer (0.1% SDS, 25 mM Tris, and 250 mM glycine). Samples were transferred onto a 0.45 μm PVDF membrane (Thermo Fisher Scientific) for 60 min at 100 mA at 4°C. Membranes were blocked in 0.2% BSA and 2% milk diluted in TBS with 0.2% Tween 20. Dysferlin detection was achieved with a 1:1,000 dilution of rabbit-anti-dysferlin (Abcam, Cambridge, UK). GAPDH was probed with a 1:10,000 dilution of rabbit-anti-GAPDH (Abcam) in blocking solution. Rabbit antibodies were probed with either a 1:5,000 dilution of goat-anti-rabbit immunoglobulin G (IgG) horseradish peroxidase (HRP) secondary antibody (Thermo Fisher Scientific) or a dilution of IRDye secondary antibodies (LI-COR Biosciences, Lincoln, NE). Blots were developed in Clarify Western ECL Substrate according to the manufacturer’s protocol (Bio-Rad) and imaged using the ChemiDoc Touch Imaging System (Bio-Rad) or the Odyssey CLx (LI-COR Biosciences).
Immunohistochemistry
Muscles were harvested and snap frozen in OCT in liquid nitrogen. Serial 12-μm cryostat sections were obtained throughout the muscles. Sections were fixed in 4% paraformaldehyde (PFA) for 10 min and washed 3 times with PBS for 1–2 min. Sections were blocked in 10% donkey serum in PBS for 1 hr. Primary antibodies were prepared in block solution and incubated overnight at 4°C in a humid chamber. The next day, slides were washed 3 times with PBS and stained with 2nd antibody for 30 min at room temperature. Slides were washed with PBS 3 times and visualized. Dysferlin was detected with the rabbit NCL-Hamlet antibody (1:500, Leica Biosystems, Buffalo Grove, IL), and laminin was detected with a rat α-laminin antibody (1:1,000, Sigma, St. Louis, MO). Goat α-rabbit conjugated to Alexa 488 (1:1,000, Life Technologies, Carlsbad, CA) and α-rat conjugated to Alexa 594 (1:1,000, Life Technologies) were used as secondary antibodies.
ELISA for Follistatin
Follistatin levels were measured using a follistatin ELISA kit (R&D Systems, London, UK) following instructions provided by the manufacturer. A standard curve was constructed using the provided follistatin standards. Protein extract samples from each muscle group were diluted 1:10 in Calibrator Diluent RD5-21. Both standards and samples were pipetted into wells pre-coated with immobilized antibody, followed by incubation with a follistatin-specific enzyme-linked monoclonal antibody. Substrate solution was then added to each well, with color development proportional to the amount of follistatin bound. Plates were read with a Tecan Infinite 200Pro microplate reader (Tecan Group, Mannedorf, Switzerland) at 450 nm, with its reference at 570 nm.
PCR
Presence of therapeutic plasmid in muscle was detected by PCR. Genomic DNA was extracted from muscles using the Wizard Genomic DNA Purification Kit according to the manufacturer’s instructions (Promega, Madison, WI). Plasmid amplicon was generated from 200 ng genomic DNA (3 min at 98°C; 20 s at 98°C, 25 s at 63°C, and 60 s at 72°C, 30 cycles; 2 min at 72°C), with the forward primer binding in the firefly luciferase gene (5′- TCTCATCTACCTCCCGGTTTT-3′) and the reverse primer binding in the rabbit β-globin UTR region (5′-TTTTGGCAGAGGGAAAAAGA-3′) using 2xQ5 MasterMix (New England Biolabs, Ipswich, MA).
Evan’s Blue Dye Procedure
To evaluate the membrane permeability of muscle, mice were intraperitoneally injected with Evan’s blue dye (Sigma; 100 μL 1% Evan’s blue dye in PBS per 10 g body weight). The following day, muscles were flash frozen and ground with a mortar and pestle. Following an overnight incubation in formamide, Evan’s blue dye was quantified in the supernatant by measuring light emission at 630 nm.39
Statistical Analysis
To determine statistical significance for two groups, comparisons were made using a Student’s t test; for more than 2 groups, the ANOVA test was used. The F test was used to analyze the variance variability of data. Statistical analyses were performed using GraphPad Prism v.7 (GraphPad Software, La Jolla, CA). A p value < 0.05 was considered significant.
Author Contributions
J.M., C.P., H.d.B., and M.B. designed and performed the experiments, interpreted results, and contributed to preparing the manuscript; M.P.C. designed the experiments, interpreted results, obtained funding, and drafted the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
We thank Rafael Contreras Lopez for initial work on the delivery method in the Calos lab and Drs. Christine Wooddell and Julia Hegge of the former Wolff lab for helpful discussions and detailed instructions on the injection method. This work was supported by grants to M.P.C. from the Muscular Dystrophy Association, Jain Foundation, Stanford Discovery Innovation Fund, and James Kanagy Fund.
References
- 1.Mercuri E., Muntoni F. Muscular dystrophies. Lancet. 2013;381:845–860. doi: 10.1016/S0140-6736(12)61897-2. [DOI] [PubMed] [Google Scholar]
- 2.Guiraud S., Aartsma-Rus A., Vieira N.M., Davies K.E., van Ommen G.J., Kunkel L.M. The pathogenesis and therapy of muscular dystrophies. Annu. Rev. Genomics Hum. Genet. 2015;16:281–308. doi: 10.1146/annurev-genom-090314-025003. [DOI] [PubMed] [Google Scholar]
- 3.Liu J., Aoki M., Illa I., Wu C., Fardeau M., Angelini C., Serrano C., Urtizberea J.A., Hentati F., Hamida M.B. Dysferlin, a novel skeletal muscle gene, is mutated in Miyoshi myopathy and limb girdle muscular dystrophy. Nat. Genet. 1998;20:31–36. doi: 10.1038/1682. [DOI] [PubMed] [Google Scholar]
- 4.Bashir R., Britton S., Strachan T., Keers S., Vafiadaki E., Lako M., Richard I., Marchand S., Bourg N., Argov Z. A gene related to Caenorhabditis elegans spermatogenesis factor fer-1 is mutated in limb-girdle muscular dystrophy type 2B. Nat. Genet. 1998;20:37–42. doi: 10.1038/1689. [DOI] [PubMed] [Google Scholar]
- 5.Bansal D., Miyake K., Vogel S.S., Groh S., Chen C.C., Williamson R., McNeil P.L., Campbell K.P. Defective membrane repair in dysferlin-deficient muscular dystrophy. Nature. 2003;423:168–172. doi: 10.1038/nature01573. [DOI] [PubMed] [Google Scholar]
- 6.Ho M., Post C.M., Donahue L.R., Lidov H.G.W., Bronson R.T., Goolsby H., Watkins S.C., Cox G.A., Brown R.H., Jr. Disruption of muscle membrane and phenotype divergence in two novel mouse models of dysferlin deficiency. Hum. Mol. Genet. 2004;13:1999–2010. doi: 10.1093/hmg/ddh212. [DOI] [PubMed] [Google Scholar]
- 7.Maguire K.K., Lim L., Speedy S., Rando T.A. Assessment of disease activity in muscular dystrophies by noninvasive imaging. J. Clin. Invest. 2013;123:2298–2305. doi: 10.1172/JCI68458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lostal W., Bartoli M., Bourg N., Roudaut C., Bentaïb A., Miyake K., Guerchet N., Fougerousse F., McNeil P., Richard I. Efficient recovery of dysferlin deficiency by dual adeno-associated vector-mediated gene transfer. Hum. Mol. Genet. 2010;19:1897–1907. doi: 10.1093/hmg/ddq065. [DOI] [PubMed] [Google Scholar]
- 9.Pryadkina M., Lostal W., Bourg N., Charton K., Roudaut C., Hirsch M.L., Richard I. A comparison of AAV strategies distinguishes overlapping vectors for efficient systemic delivery of the 6.2 kb Dysferlin coding sequence. Mol. Ther. Methods Clin. Dev. 2015;2:15009. doi: 10.1038/mtm.2015.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sondergaard P.C., Griffin D.A., Pozsgai E.R., Johnson R.W., Grose W.E., Heller K.N., Shontz K.M., Montgomery C.L., Liu J., Clark K.R. AAV.dysferlin overlap vectors restore function in dysferlinopathy animal models. Ann. Clin. Transl. Neurol. 2015;2:256–270. doi: 10.1002/acn3.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Llanga T., Nagy N., Conatser L., Dial C., Sutton R.B., Hirsch M.L. Structure-based designed nano-dysferlin that significantly improves dysferlinopathy in BLA/J mice. Mol. Ther. 2017;25:2150–2162. doi: 10.1016/j.ymthe.2017.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Louis Jeune V., Joergensen J.A., Hajjar R.J., Weber T. Pre-existing anti-adeno-associated virus antibodies as a challenge in AAV gene therapy. Hum. Gene Ther. Methods. 2013;24:59–67. doi: 10.1089/hgtb.2012.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertoni C., Jarrahian S., Wheeler T.M., Li Y., Olivares E.C., Calos M.P., Rando T.A. Enhancement of plasmid-mediated gene therapy for muscular dystrophy by directed plasmid integration. Proc. Natl. Acad. Sci. USA. 2006;103:419–424. doi: 10.1073/pnas.0504505102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolff J.A., Malone R.W., Williams P., Chong W., Acsadi G., Jani A., Felgner P.L. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 15.Hagstrom J.E., Hegge J., Zhang G., Noble M., Budker V., Lewis D.L., Herweijer H., Wolff J.A. A facile nonviral method for delivering genes and siRNAs to skeletal muscle of mammalian limbs. Mol. Ther. 2004;10:386–398. doi: 10.1016/j.ymthe.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 16.Zhang G., Wooddell C.I., Hegge J.O., Griffin J.B., Huss T., Braun S., Wolff J.A. Functional efficacy of dystrophin expression from plasmids delivered to mdx mice by hydrodynamic limb vein injection. Hum. Gene Ther. 2010;21:221–237. doi: 10.1089/hum.2009.133. [DOI] [PubMed] [Google Scholar]
- 17.Hegge J.O., Wooddell C.I., Zhang G., Hagstrom J.E., Braun S., Huss T., Sebestyén M.G., Emborg M.E., Wolff J.A. Evaluation of hydrodynamic limb vein injections in nonhuman primates. Hum. Gene Ther. 2010;21:829–842. doi: 10.1089/hum.2009.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wooddell C.I., Hegge J.O., Zhang G., Sebestyén M.G., Noble M., Griffin J.B., Pfannes L.V., Herweijer H., Hagstrom J.E., Braun S. Dose response in rodents and nonhuman primates after hydrodynamic limb vein delivery of naked plasmid DNA. Hum. Gene Ther. 2011;22:889–903. doi: 10.1089/hum.2010.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan Z., Kocis K., Valley R., Howard J.F., Chopra M., An H., Lin W., Muenzer J., Powers W., Powers W. Safety and feasibility of high-pressure transvenous limb perfusion with 0.9% saline in human muscular dystrophy. Mol. Ther. 2012;20:456–461. doi: 10.1038/mt.2011.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan Z., Kocis K., Valley R., Howard J.F., Jr., Chopra M., Chen Y., An H., Lin W., Muenzer J., Powers W. High-pressure transvenous perfusion of the upper extremity in human muscular dystrophy: a safety study with 0.9% saline. Hum. Gene Ther. 2015;26:614–621. doi: 10.1089/hum.2015.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee S.J., McPherron A.C. Regulation of myostatin activity and muscle growth. Proc. Natl. Acad. Sci. USA. 2001;98:9306–9311. doi: 10.1073/pnas.151270098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodino-Klapac L.R., Janssen P.M.L., Shontz K.M., Canan B., Montgomery C.L., Griffin D., Heller K., Schmelzer L., Handy C., Clark K.R. Micro-dystrophin and follistatin co-delivery restores muscle function in aged DMD model. Hum. Mol. Genet. 2013;22:4929–4937. doi: 10.1093/hmg/ddt342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee Y.S., Lehar A., Sebald S., Liu M., Swaggart K.A., Talbot C.C., Jr., Pytel P., Barton E.R., McNally E.M., Lee S.J. Muscle hypertrophy induced by myostatin inhibition accelerates degeneration in dysferlinopathy. Hum. Mol. Genet. 2015;24:5711–5719. doi: 10.1093/hmg/ddv288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turan S., Farruggio A.P., Srifa W., Day J.W., Calos M.P. Precise correction of disease mutations in induced pluripotent stem cells derived from patients with limb girdle muscular dystrophy. Mol. Ther. 2016;24:685–696. doi: 10.1038/mt.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyazaki J., Takaki S., Araki K., Tashiro F., Tominaga A., Takatsu K., Yamamura K. Expression vector system based on the chicken β-actin promoter directs efficient production of interleukin-5. Gene. 1989;79:269–277. doi: 10.1016/0378-1119(89)90209-6. [DOI] [PubMed] [Google Scholar]
- 26.Niwa H., Yamamura K., Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 27.Chung J.H., Whiteley M., Felsenfeld G. A 5′ element of the chicken β-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 28.Zhao C., Farruggio A.P., Bjornson C.R.R., Chavez C.L., Geisinger J.M., Neal T.L., Karow M., Calos M.P. Recombinase-mediated reprogramming and dystrophin gene addition in mdx mouse induced pluripotent stem cells. PLoS One. 2014;9:e96279. doi: 10.1371/journal.pone.0096279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pichavant C., Bjornson C.R.R., Gallagher T.J., Neal T., du Bois H., Bhakta M., Calos M.P. Three novel immune-deficient mouse models of muscular dystrophy. PLoS Curr. Musc. Dystr. 2017 Published online September 1, 2017. [Google Scholar]
- 30.Calos M.P. Phage integrases for genome editing. In: Cathomen T., Hirsch M., Porteus M., editors. Genome Editing: The Next Step in Gene Therapy. Springer; 2016. pp. 81–91. [Google Scholar]
- 31.Calos M.P. Genome editing techniques and their therapeutic applications. Clin. Pharmacol. Ther. 2017;101:42–51. doi: 10.1002/cpt.542. [DOI] [PubMed] [Google Scholar]
- 32.Arnett A.L.H., Konieczny P., Ramos J.N., Hall J., Odom G., Yablonka-Reuveni Z., Chamberlain J.R., Chamberlain J.S. Adeno-associated viral (AAV) vectors do not efficiently target muscle satellite cells. Mol. Ther. Methods Clin. Dev. 2014;1:14038. doi: 10.1038/mtm.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee S.-J. Quadrupling muscle mass in mice by targeting TGF-β signaling pathways. PLoS One. 2007;2:e789. doi: 10.1371/journal.pone.0000789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mendell J.R., Sahenk Z., Malik V., Gomez A.M., Flanigan K.M., Lowes L.P., Alfano L.N., Berry K., Meadows E., Lewis S. A phase 1/2a follistatin gene therapy trial for becker muscular dystrophy. Mol. Ther. 2015;23:192–201. doi: 10.1038/mt.2014.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roudaut C., Le Roy F., Suel L., Poupiot J., Charton K., Bartoli M., Richard I. Restriction of calpain3 expression to the skeletal muscle prevents cardiac toxicity and corrects pathology in a murine model of limb-girdle muscular dystrophy. Circulation. 2013;128:1094–1104. doi: 10.1161/CIRCULATIONAHA.113.001340. [DOI] [PubMed] [Google Scholar]
- 36.Kamimura K., Zhang G., Liu D. Image-guided, intravascular hydrodynamic gene delivery to skeletal muscle in pigs. Mol. Ther. 2010;18:93–100. doi: 10.1038/mt.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evesson F.J., Peat R.A., Lek A., Brilot F., Lo H.P., Dale R.C., Parton R.G., North K.N., Cooper S.T. Reduced plasma membrane expression of dysferlin mutants is attributed to accelerated endocytosis via a syntaxin-4-associated pathway. J. Biol. Chem. 2010;285:28529–28539. doi: 10.1074/jbc.M110.111120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pearson T., Shultz L.D., Miller D., King M., Laning J., Fodor W., Cuthbert A., Burzenski L., Gott B., Lyons B. Non-obese diabetic-recombination activating gene-1 (NOD-Rag1 null) interleukin (IL)-2 receptor common gamma chain (IL2r gamma null) null mice: a radioresistant model for human lymphohaematopoietic engraftment. Clin. Exp. Immunol. 2008;154:270–284. doi: 10.1111/j.1365-2249.2008.03753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wooddell C.I., Radley-Crabb H.G., Griffin J.B., Zhang G. Myofiber damage evaluation by Evans blue dye injection. Curr. Protoc. Mouse Biol. 2011;1:463–488. doi: 10.1002/9780470942390.mo110141. [DOI] [PubMed] [Google Scholar]