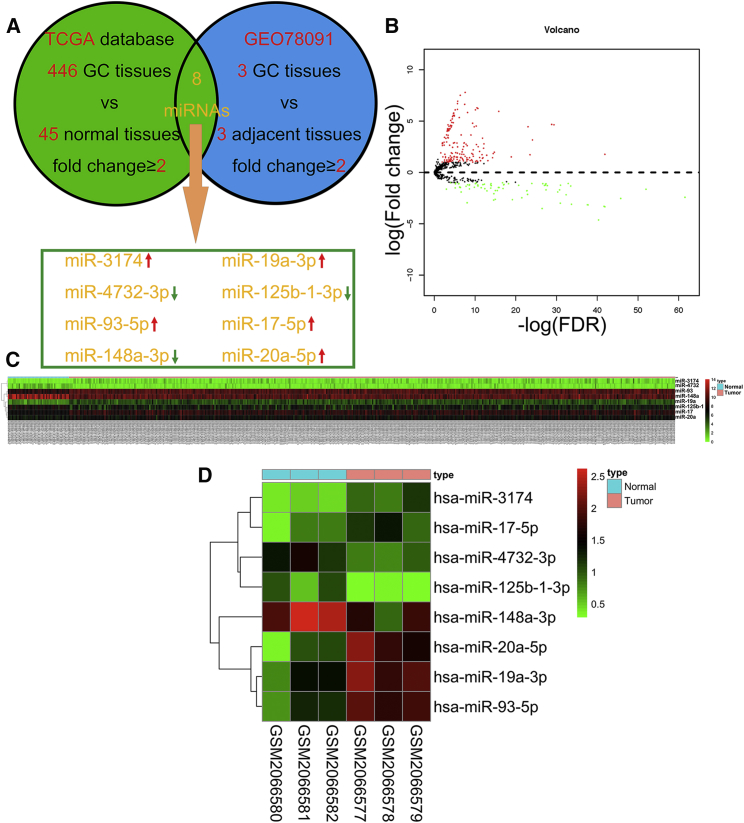
Figure 1.
Identifying Differentially Expressed miRNAs by Using TCGA and the GEO Database
(A) Bioinformatics analysis flow diagram for the screening for differentially expressed miRNAs in GC on the basis of TCGA and GEO databases. Eight miRNAs were significantly differentially expressed in GC tissues compared with normal tissues with a fold change ≥2 and p < 0.05. (B) Volcano plot of the miRNA matrix generated according to TCGA statistics from 491 GC patients. Red puncta represent highly expressed miRNAs in GC tissues in contrast to normal tissues with a fold change ≥2 and p < 0.05. Green puncta represent downregulated miRNAs in GC with fold change ≥2 and p < 0.05. (C) A miRNA expression heatmap for 446 GC patients and 45 healthy controls from TCGA was generated by using R with the pheatmap package. (D) miRNA expression heatmap for three GC tissues and three paired adjacent tissues from the GEO78091 dataset, generated by using R with the pheatmap package.