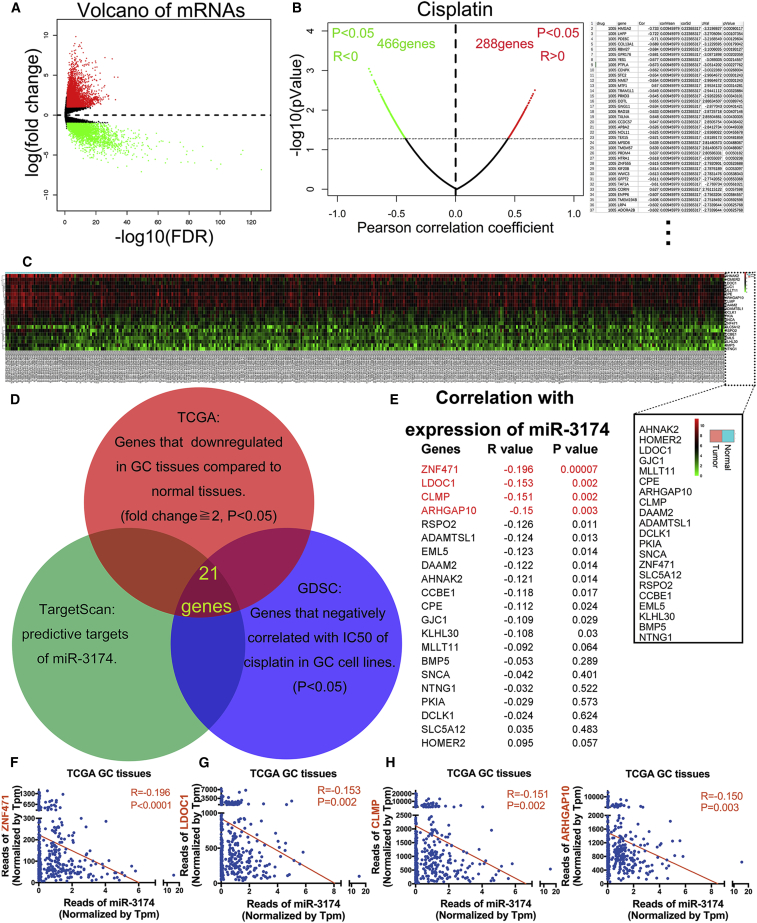
Figure 6.
Bioinformatics Analysis to Identify Potential Downstream Targets of miR-3174
(A) Volcano plot for the mRNA matrix generated on the basis of TCGA statistics from 407 patients. (B) Left panel, candidate genes potentially involved in CDDP sensitivity and resistance were screened according to the GDSC database. Right panel, 36 genes whose expression was negatively correlated with the IC50 values for CDDP in 28 different GC cell lines are listed. (C) mRNA expression heatmap showing 21 genes whose expression was not only downregulated in 375 GC tissues compared with 32 normal gastric tissues on the basis of TCGA database but also significantly negatively correlated with the IC50 values for CDDP in 28 different GC cell lines. In addition, these genes were also identified as potential targets of miR-3174 based on the TargetScan. The symbols for the 21 genes are shown in the black square. (D) Schematic diagram detailing the exploration of miR-3174 downstream targets. (E) Correlations between the expression levels of miR-3174 and these 21 genes were calculated with Pearson correlation analysis in 404 patients from TCGA database, and the gene name, R value, and p value are shown. The first four genes, ZNF471, LODC1, CLMP, and ARHGAP10, with p < 0.01 were selected for further validation (red font). (F–I) A negative correlation was found between miR-3174 and ZNF471 (F), LODC1 (G), CLMP (H), and ARHGAP10 (I) according to TCGA statistics is displayed. Graph represents mean ± SEM; *p < 0.05, **p < 0.01, and ***p < 0.001.