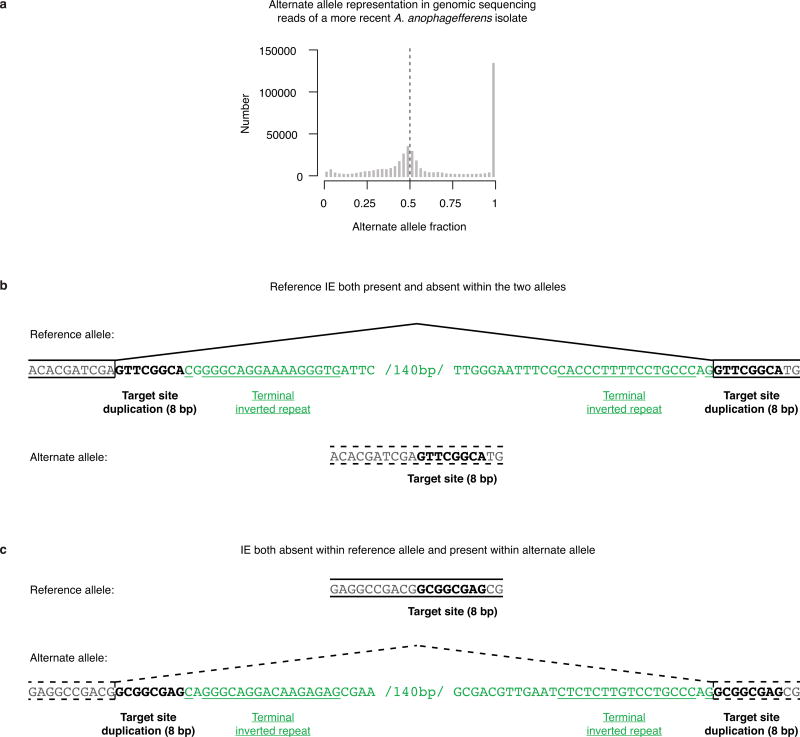
Extended Data Figure 6. Diploid genomic sequence variation in a more recent isolate of A. anophagefferens.
a, Calling of sequence variation from genomic sequencing reads without an assumption of ploidy reveals a peak at alternate allele fraction of approximately 0.5. The most likely scenario is that this A. anophagefferens isolate has a diploid genome. It is not physically plausible for it to have higher ploidy because that amount of chromatin could not fit into its extremely compact nucleus12. b, An example reference IE is present within one allele and absent within the alternate allele. The locus is displayed as in Fig. 3a. The reference IE is located in an annotated protein-coding gene with a 200 bp RNA sequencing-validated intron in the reference isolate. The alternate allele is likely exonic without an intron (broken lines), so that it encodes the same amino acid sequence. The TSD within the reference allele is 8 bp, immediately flanking the IE TIRs. c, An example IE not found within the reference allele is present within the alternate allele. The locus is displayed as in Fig. 3a. The alternate IE is within an annotated protein-coding gene with a predicted 200 bp intron (broken lines). If the predicted intron is indeed spliced out of the RNA, then the alternate allele encodes the same amino acid sequence. The TSD within the alternate allele is 8 bp, immediately flanking the IE TIRs.