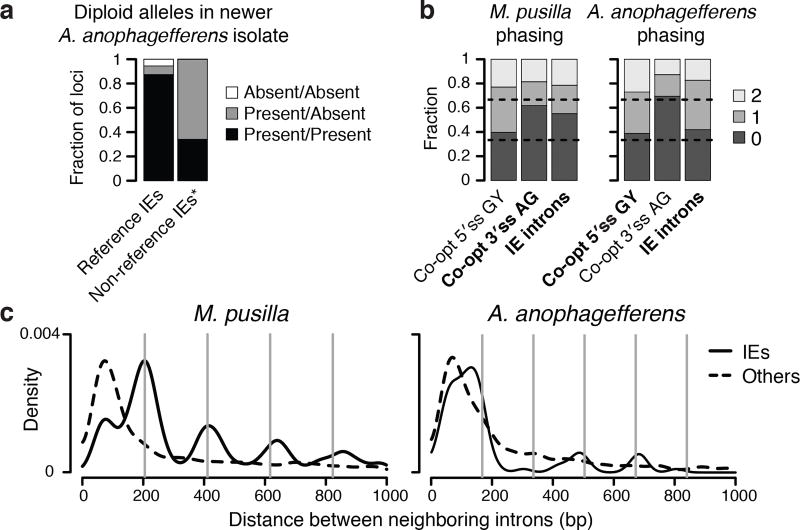
Figure 4. IE dynamics and genomic implications.
a, Presence-absence variation in a newer isolate of A. anophagefferens. *Non-reference IEs identified cannot be absent/absent. b, Sequences that can be co-opted to construct splice sites are biased with respect to codon phasing. For M. pusilla, IE introns should be biased by availability of AG sequences that can be co-opted as 3′ splice sites (3′ss). For A. anophagefferens, IE introns should be biased by availability of GY (Y is C or T) sequences that can be co-opted for 5′ splice sites (5′ss). IE introns indeed have phase biases more similar to the respectively co-opted sequence (bold). c, Nearby IE insertions generate nucleosome-sized segments. Distances between neighboring IE introns (solid) and between other neighboring introns (broken) are displayed as kernel density estimates. Nucleosome repeat lengths12 of 206 bp for M. pusilla and 168 bp for A. anophagefferens show the expected sizes of integer numbers of nucleosomes (vertical lines).