Abstract
The purpose of this study was to identify the roles of non-homologous end-joining (NHEJ) or homologous recombination (HR) pathways in repairing DNA double-strand breaks (DSBs) induced by exposure to high-energy protons and carbon ions (C ions) versus gamma rays in Chinese hamster cells. Two Chinese hamster cell lines, ovary AA8 and lung fibroblast V79, as well as various mutant sublines lacking DNA-PKcs (V3), X-ray repair cross-complementing protein-4 [XRCC4 (XR1), XRCC3 (irs1SF) and XRCC2 (irs1)] were exposed to gamma rays (137Cs), protons (200 MeV; 2.2 keV/μm) and C ions (290 MeV; 50 keV/μm). V3 and XR1 cells lack the NHEJ pathway, whereas irs1 and irs1SF cells lack the HR pathway. After each exposure, survival was measured using a clonogenic survival assay, in situ DSB induction was evaluated by immunocytochemical analysis of histone H2AX phosphorylation at serine 139 (γ-H2AX foci) and chromosome aberrations were examined using solid staining. The findings from this study showed that clonogenic survival clearly depended on the NHEJ and HR pathway statuses, and that the DNA-PKcs−/− cells (V3) were the most sensitive to all radiation types. While protons and γ rays yielded almost the same biological effects, C-ion exposure greatly enhanced the sensitivity of wild-type and HR-deficient cells. However, no significant enhancement of sensitivity in cell killing was seen after C-ion irradiation of NHEJ deficient cells. Decreases in the number of γ-H2AX foci after irradiation occurred more slowly in the NHEJ deficient cells. In particular, V3 cells had the highest number of residual γ-H2AX foci at 24 h after C-ion irradiation. Chromosomal aberrations were significantly higher in both the NHEJ- and HR-deficient cell lines than in wild-type cell lines in response to all radiation types. Protons and gamma rays induced the same aberration levels in each cell line, whereas C ions introduced higher but not significantly different aberration levels. Our results suggest that the NHEJ pathway plays an important role in repairing DSBs induced by both clinical proton and C-ion beams. Furthermore, in C ions the HR pathway appears to be involved in the repair of DSBs to a greater extent compared to gamma rays and protons.
INTRODUCTION
Proton and carbon-ion (C-ion) beams have been used to treat solid cancers, and the numbers of treatment facilities and patients undergoing proton and C-ion therapy are increasing rapidly as a result of the excellent localized dose distributions and preservation of surrounding normal tissues offered by these technologies.
To further improve the efficacy of these particles beam radiotherapies, it is important to clarify the molecular mechanisms of both the tumor and normal tissue responses to these particle beams, since these may facilitate particle-specific radiosensitization. The biological characteristics of particle beams and photons have been analyzed and compared using different end points (1–6). Furthermore, it has been reported that the biological effectiveness of particle beams might vary depending on the biological end points as well as the tissues targeted or cell lines irradiated (3, 4). In our previous study, we observed that proton beams induced greater rates of apoptosis than did photons, and the apoptosis induction ratios were significantly higher than the relative biological effectiveness (RBE) values calculated at 10% survival of a clonogenic survival assay (4). In addition, although C-ion beams have been shown to yield greater RBE values than protons, the characteristics of DNA lesions and their repair mechanisms are not fully understood (7).
It is known that DNA double-strand breaks (DSBs) are a lethal type of damage induced by ionizing radiation, and the majority of DSBs induced are repaired either through the non-homologous end-joining (NHEJ) or homologous recombination (HR) pathway. Ku70/80 proteins, which are abundant in cells, instantly recognize DSB ends because of their high affinity for these DNA ends. In the NHEJ pathway, after Ku70/80 binds to DSB ends, DNA-PKcs is recruited to the damaged sites and the XRCC4-DNA ligase IV complex subsequently re-ligates the two DSB ends (8). Recently published studies have clarified that Ku70/80 binding protects the DNA ends from unnecessary resection and inhibits HR pathway initiation (9–11).
The HR pathway uses a homologous template to repair DSBs and is therefore cell cycle dependent. HR pathway initiation is mediated by the recognition of DSB ends by the Mre11, Rad50 and Nbs1 (MRN) complex and end resection by the CtBP-interacting protein, which associates with the MRN complex and BRCA2. Next, single-stranded DNA tails are stabilized by the RPA protein and subsequently replaced by RAD51 with the help of recombination mediators (9, 12, 13).
Several published studies have addressed the different contributions of the NHEJ and HR pathways to DSB repair according to the complexity of the DSBs and the cell cycle phase in which the cells are irradiated (14–17). In addition, it was recently reported that high-linear energy transfer (LET) particle beams induced more complex and more fragmented DNA lesions, and the involvement of HR in the repair of these lesions was greater than that observed for low-LET photon irradiated lesions (17–20). However, this phenomenon has not been fully investigated because few studies have addressed the characteristics of particle-beam-induced DNA lesions and the associated repair pathways. Furthermore, the contributions of the NHEJ and HR pathways to the repair of clinical proton and C-ion beams and photon-induced DSBs have not been clarified. Therefore, in this study we investigated the contributions of the NHEJ and HR pathways to the repair of proton- and C-ion-induced DNA lesions. We exposed Chinese hamster cells defective in either the NHEJ or HR pathway to γ rays, protons and C ions and examined radiosensitivity, γ-H2AX foci formation kinetics and genome stability maintenance.
MATERIALS AND METHODS
Cell Lines and Culture Conditions
The wild-type Chinese hamster ovary (CHO) cell line AA8 (21) and the Chinese hamster lung fibroblast cell line V79 (22), as well as various mutant sublines defective in either the NHEJ or HR pathway were used in this study. Specifically, the DNA-PKcs−/− cell line (V3) (16) and XRCC4−/− cell line (XR1) (23) used in this study lacked the NHEJ pathway, whereas the XRCC3−/− cell line (irs1SF) (24) and XRCC2−/− cell line (irs1) (25) used in this study lacked the HR pathway. The irs1SF cell line was kindly provided by Dr. Takamitsu Kato at Colorado State University (Ft. Collins, CO).
Cells were grown in minimum essential media (Sigma-Aldrich Japan, Tokyo, Japan) supplemented with 100 μg/mL streptomycin, 100 U/mL penicillin (Sigma-Aldrich) and 10% fetal bovine serum (Sigma-Aldrich). For cell transfer, the cells were rinsed with Ca2+ and Mg2+-free phosphate buffered saline (PBS−) (Sigma-Aldrich) and dispersed in 0.25% trypsin solution containing 0.5 mM ethylenedi-aminetetraacetate (EDTA) (Sigma-Aldrich). The cells were maintained at 37°C in a humidified incubator under 5% CO2. The doubling times of all cell lines ranged from 16–20 h.
The number of cells was determined using a T10 Automated Cell Counter (Bio-Rad Laboratories, Tokyo, Japan). For radiation exposure in a subconfluent state, the cells were seeded into either T75 or T25 flasks (NUNC; Thermo Fisher Scientific, Roskilde, Denmark) or onto glass cover slips (Matsunami Glass Ind. Ltd., Osaka, Japan) in 6-well plates at a densities of 1 ×106, 1 ×105 and 2 ×104, respectively and subsequently, then incubated for at least 24 h for clonogenic survival assay and chromosome aberration analysis, and 48 h for γ-H2AX foci assay under the conditions described above.
Irradiation
Dosimetry
For 137Cs γ-ray irradiation, the GammaCell® 40 (Atomic Energy of Canada, Ontario, Canada) was used. The dose rate was 0.76 Gy/min, which was calculated from the decay curve of the 137Cs source. For proton beam irradiation, 200 MeV proton beams were generated by the synchrotron at the Proton Medical Research Center (PMRC) at the University of Tsukuba, (Tsukuba, Japan). Proton dosimetry was measured as previously described (26, 27). Based on the 200 MeV proton beam dose distribution, the absorbed dose was measured at the middle of the 6 cm wide spread-out Bragg peak (SOBP) using a dosimeter (0.2 cc C-110 Farmer Chamber; Applied Engineering Inc., Tokyo, Japan). The estimated energy spread and dose-average LET values at the mid-SOBP were 0–60 MeV and 2.2 keV/μm, respectively (27, 28).
For C-ion beam exposure, 290 MeV/n C-ion beams were generated by the synchrotron at the Heavy Ion Medical Accelerator in Chiba (HIMAC) (National Institute of Radiological Sciences, Japan). Details concerning the beam characteristics, biological irradiation procedures and C-ion beam dosimetry have been previously described (29, 30). The experimental dose distributions were measured at the middle of a 6 cm SOBP of C ions, and the energy spread and the dose-average LET values were approximately 0–160 MeV and 50 keV/μm, respectively (7).
Irradiation conditions and selected doses
Proton and C-ion irradiations were performed at a dose rate of approximately 3 Gy/min at the middle of the 6 cm SOBP in a 10 × 10 cm field. The dose was adjusted according to specific parameters (e.g., temperature and atmospheric pressure) in each experiment. Each exposure was performed at room temperature (25.5–26.0°C).
For clonogenic survival and chromosomal analyses, cells growing exponentially in T25 or T75 flasks and for detection of γ-H2AX foci cells growing on glass cover slips in 6-well plates were placed at the middle of the proton and C-ion SOBPs and irradiated from the bottom. Gamma-ray irradiation was performed from both the top and bottom.
The dose points selected for the wild-type cell colony formation assays were 0, 1, 2, 4 and 8 Gy, and those selected for the repair-deficient cell colony formation assays were 0, 0.5, 1, 2 and 4 Gy. In addition, the dose point selected for γ-H2AX foci detection was 2 Gy, since this is the standard dose per fraction used in clinical practice. As for chromosomal aberration, we selected 1 Gy to compare our results with other published studies, since CHO and V79 cells are well studied with 1 Gy or less for chromosomal aberration (31).
Clonogenic Survival Assay
Immediately after each exposure, the cells were dispersed with 0.25% trypsin-EDTA and seeded onto 60 mm culture dishes at appropriate densities. Colonies of wild-type and repair-deficient cell lines were fixed and stained after day 10 and 14 of incubation, respectively. Five replicate dishes were seeded for each dose point, and colonies containing more than 50 cells were scored as survivors.
The resulting survival data were fitted according to the linear-quadratic (LQ) model using the GraphPad Prism software (GraphPad Software Inc., San Diego, CA). From these curves, the doses corresponding to 10% survival (D10), RBE0.1 values, survival fractions at 2 Gy (SF2), and α and β values were calculated and used to compare the effects of γ rays, proton beams and C-ion beams.
Immunocytochemical Staining of γ-H2AX
After each exposure, the cells were fixed in 4% formaldehyde (Wako Pure Chemical Industries, Osaka, Japan) for 15 min at 0.5, 3, 5, 7, 12 and 24 h time points. The fixed cells were immunocytochemically stained with human monoclonal Anti-phospho-Histone H2AX (Ser139) Antibody (EMD Millipore, Darmstadt, Germany) according to the manufacturer’s protocol with minor modifications. In brief, the fixed cells were washed in PBS− and permeabilized in 0.5% Triton X-100 (Wako Pure Chemical Industries). Next, the cells were rinsed again in PBS− and blocked with 3% bovine serum albumin (Wako Pure Chemical Industries) in PBS− with 0.5% Tween-20 (Wako Pure Chemical Industries) for 30 min at room temperature in a humidified chamber. The cells were sequentially incubated for 1.5 h with a 1:1,000 dilution of human monoclonal Anti-phospho-Histone H2AX (Ser139) antibody in blocking buffer and for 1 h with a 1:1,000 dilution of Alexa Fluor® 488-conjugated donkey anti-mouse immunoglobulin G (H+L) secondary antibody (Invitrogen/Molecular Probes, Eugene, OR) in blocking buffer. The cells were subsequently counterstained with a 2 μg/mL solution of 4′,6-diamidino-2-phenyl-indole dihydrochloride n-hydrate (DAPI; Wako Pure Chemical Industries) and finally mounted with DAKO fluorescent mounting medium (DAKO North America, Carpinteria, CA) on slide glasses.
The resulting fluorescent images were captured using a fluorescence microscope (Biozero BZ-8000 Keyence; Tokyo, Japan), and the accompanying software was used to obtain Z-stacking images consisting of 4–6 images with 0.7 μm thickness in each nucleus to avoid possible overlapping of foci. From the obtained images, at least 50 nuclei per experiment and a total of more than 150 nuclei with distinct foci were selected for analysis. The number of foci was counted using “foci counter” software as previously described (5).
Chromosomal Aberration Analysis
To detect radiation-induced chromosome aberrations, colcemid (Irvine Scientific, Santa Ana, CA) was added to the culture media at a final concentration of 0.1 μg/mL at 3 and 16 h after irradiation and incubated for an additional 4 h. After incubation with colcemid, the cells were dispersed and incubated in a hypotonic (75 mM) KCl (Wako Pure Chemical Industries) solution. Next, the cells were fixed in a 3:1 methanol:acetic acid solution (Wako Pure Chemical Industries) and air dried, and chromosome spreads were made as previously described (32). After Giemsa staining, the chromosomal aberrations were scored according to the conventions outlined by Savage (33).
Statistical Analysis
The experimental values are expressed as means of at least 3 independent experiments, and the error bars indicate the standard deviation (SD) calculated for each data point. Significant differences between data were assessed with Student’s t test. The probability value of P < 0.05 was considered significant for all selected dose and time points. In addition, confidence interval (CI) estimation was performed for the colony survival assay, in which the RBE, α and β values were calculated.
RESULTS
Colony Formation Assay
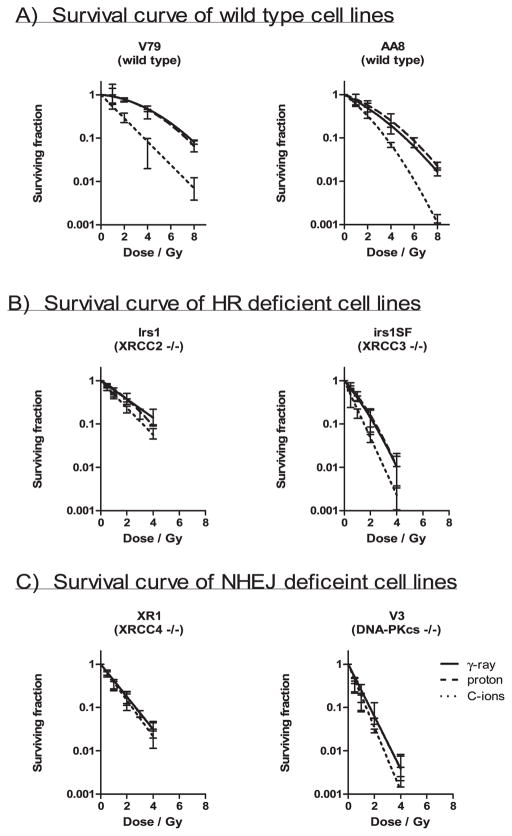
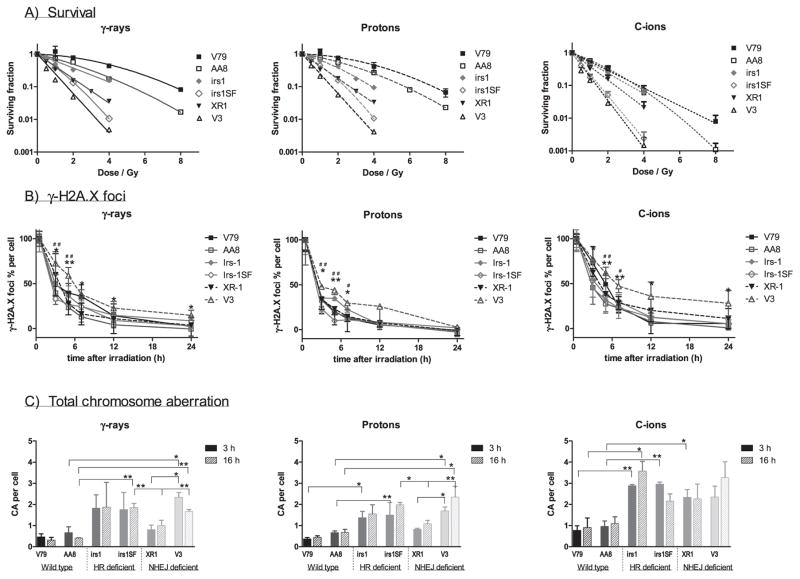
The results of the colony formation assays are shown in Fig. 1. Although shoulders were observed on the γ rays and proton beam-induced survival curves for the wild-type V79 and AA8 cells and C-ion beam-induced survival curve of wild-type AA8 cell, these were not observed on the corresponding survival curves for the repair-deficient cell lines or C-ion beam-induced survival curves for wild-type V79 (Fig. 1). However, as the calculated errors of the β parameters were the same or higher than the mean value, as shown in Table 1, the shape of the curves with the variable β parameters may be inconsistent. The wild-type cells were most resistant, followed by the HR-deficient irs1, NHEJ-deficient XR1, HR-deficient irs1SF and NHEJ-deficient V3 cells to all radiation types examined (Fig. 4A). The cells responded to γ rays and proton beams in a nearly identical manner; however, the wild-type and HR-deficient cell lines were more strongly sensitized to C ions than the NHEJ-deficient cell lines. In particular, the cytotoxic effect of C ions on XR1 cells, which lack XRCC4, was smaller than that observed in the other cell lines (Figs. 1 and 4A).
FIG. 1.
Colony survival assay. Clonogenic survival curves are shown for Chinese hamster cell lines, wild-type V79 and AA8 (panel A), HR-deficient irs1 and irs1SF (panel B). NHEJ-deficient XR1 and V3 (panel C) after irradiation with γ rays or 200 MeV protons and 290 MeV C ions with 6 cm SOBP. The curves were fitted by LQ model and the error bars represent SD.
TABLE 1.
Parameters of Survival Assay
| Cell line | Radiation type | α ± 95 CI (Gy−1) | β ± 95 CI (Gy−2) | SF2 ± 95 CI | D10 ± 95 CI (Gy) | RBE0.1 ± 95 CI |
|---|---|---|---|---|---|---|
| Wild-type | ||||||
| CH lung fibroblast V79 | γ rays | 0.05 ± 0.08 | 0.04 ± 0.01 | 0.78 ± 0.13 | 7.26 ± 0.37 | – |
| protons | 0.05 ± 0.18 | 0.05 ± 0.03 | 0.75 ± 0.13 | 6.68 ± 1.99 | 1.10 ± 0.25 | |
| C ions | 0.59 ± 0.31 | 0.02 ± 0.06 | 0.30 ± 0.13 | 3.49 ± 0.94 | 2.10 ± 0.47 | |
| CH ovarian AA8 | γ rays | 0.23 ± 0.15 | 0.05 ± 0.06 | 0.57 ± 0.12 | 5.11 ± 1.25 | – |
| protons | 0.25 ± 0.46 | 0.03 ± 0.07 | 0.56 ± 0.31 | 5.64 ± 1.45 | 0.91 ± 0.02 | |
| C ions | 0.51 ± 0.05 | 0.03 ± 0.03 | 0.34 ± 0.12 | 3.71 ± 0.68 | 1.37 ± 0.08 | |
| HR deficient | ||||||
| irs1 (XRCC2−/−) (subline of V79) | γ rays | 0.59 ± 0.11 | 5.85e-014 ± 1.02e-013 | 0.33 ± 0.10 | 3.88 ± 0.72 | – |
| protons | 0.38 ± 0.14 | 0.06 ± 0.05 | 0.42 ± 0.17 | 3.89 ± 0.90 | 1.00 ± 0.06 | |
| C ions | 0.84 ± 0.20 | 3.07e-014 ± 1.00e-013 | 0.22 ± 0.08 | 2.78 ± 0.65 | 1.40 ± 0.17 | |
| irs1SF (XRCC3−/−) (subline of AA8) | γ rays | 0.46 ± 0.80 | 0.43 ± 0.88 | 0.13 ± 0.15 | 2.00 ± 0.99 | – |
| protons | 0.48 ± 0.61 | 0.35 ± 0.92 | 0.15 ± 0.14 | 2.29 ± 1.31 | 0.89 ± 0.27 | |
| C ions | 1.63 ± 1.72 | 0.27 ± 0.92 | 0.05 ± 0.02 | 1.17 ± 0.31 | 1.70 ± 0.41 | |
| NHEJ deficient | ||||||
| XR1 (XRCC4−/−) (subline of AA8) | γ rays | 1.04 ± 0.51 | 3.40e-004 ± 1.15e-003 | 0.16 ± 0.14 | 2.29 ± 1.01 | – |
| protons | 1.01 ± 0.09 | 1.94e-012 ± 6.58e-012 | 0.18 ± 0.12 | 2.28 ± 0.21 | 1.00 ± 0.41 | |
| C ions | 0.92 ± 0.48 | 0.10 ± 0.35 | 0.16 ± 0.10 | 2.15 ± 1.01 | 1.07 ± 0.33 | |
| V3 (DNA-PKcs−/−) (subline of AA8) | γ rays | 1.93 ± 1.59 | 0.18 ± 0.63 | 0.05 ± 0.04 | 1.11 ± 0.47 | – |
| protons | 1.38 ± 0.13 | 0.34 ± 0.94 | 0.06 ± 0.13 | 1.40 ± 0.92 | 0.94 ± 0.61 | |
| C ions | 2.39 ± 1.07 | 5.34e-013 ± 1.56e-012 | 0.03 ± 0.006 | 1.00 ± 0.04 | 1.11 ± 0.10 | |
Notes. The α and β values and the percentage of surviving colonies after a single dose of 2 Gy radiation (SF2) were analyzed using nonlinear regression with GraphPad Prism software according to the linear-quadratic model. D10 denotes a single dose of γ rays, protons or C ions that produce a 10% survival rate. RBE0.1 indicates RBE at a 10% survival rate. A confidence interval of 95% was calculated from 3 independent experiments. CH = Chinese hamster; CI = confidence interval.
FIG. 4.
Comparison of cell lines. Panel A: Survival curves of cell lines were plotted by each radiation type; γ rays, protons and C ions. The wild-type V79 (■) and AA8 (□) cells were most resistant, followed by HR-deficient irs1 (
 ) and NHEJ-deficient XR1 (▼); HR-deficient irs1SF (
) and NHEJ-deficient XR1 (▼); HR-deficient irs1SF (
 ) and NHEJ-deficient V3 (△) were found to be the most sensitive to all radiation types examined. Panel B: Formation and dissolution of γ-H2AX foci after irradiation with γ rays, protons and C ions. Student’s t test: **P < 0.01, *P < 0.05 of V3 compared with wild-type cell; ##P < 0.01, #P < 0.05 of V3 compared with HR-deficient irs1SF cell. Panel C: Chromosome aberrations induced by 1 Gy γ ray, protons and C ions after 3 (filled column) and 16 h (gradient column) irradiations were plotted in each cell line examined after subtracting the number of 0 Gy irradiated cells. Student’s t test: **P < 0.01, *P < 0.05. Error bars represent SD.
) and NHEJ-deficient V3 (△) were found to be the most sensitive to all radiation types examined. Panel B: Formation and dissolution of γ-H2AX foci after irradiation with γ rays, protons and C ions. Student’s t test: **P < 0.01, *P < 0.05 of V3 compared with wild-type cell; ##P < 0.01, #P < 0.05 of V3 compared with HR-deficient irs1SF cell. Panel C: Chromosome aberrations induced by 1 Gy γ ray, protons and C ions after 3 (filled column) and 16 h (gradient column) irradiations were plotted in each cell line examined after subtracting the number of 0 Gy irradiated cells. Student’s t test: **P < 0.01, *P < 0.05. Error bars represent SD.
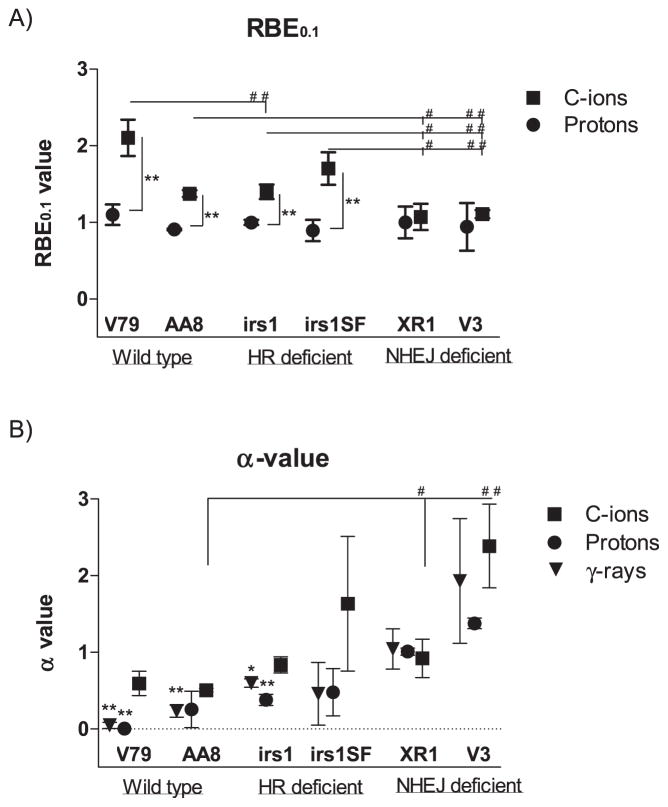
As shown in Table 1 and Fig. 2A, the RBE values at 10% survival (RBE0.1) of protons to γ rays ranged from 0.89–1.10, and no differences were observed among the wild-type, HR-deficient and NHEJ-deficient cell lines. In contrast, the RBE0.1 value of C ions to γ rays ranged from 1.07–2.10, and the RBE0.1 values of the wild-type (2.10 ± 0.47 in V79; 1.37 ± 0.08 in AA8) and HR-deficient (1.40 ± 0.17 in irs1; 1.70 ± 0.41 in irs1SF) cell lines were significantly higher than those of the NHEJ-deficient cell lines (1.07 ± 0.33 in XR1; 1.11 ± 0.10 in V3; Fig. 2A and Table 1). The 95% CI for the proton beam RBE0.1 values did not significantly exceed 1.00 in any of the examined cell lines. However, the 95% CIs of the C-ion RBE0.1 values were significantly greater than 1.00 in the wild-type and HR-deficient cell lines but not in the NHEJ-deficient cell lines (Table 1). In addition, the α values for each radiation type, which were calculated according to the LQ model, were plotted for each cell line. The α values of the protons and γ rays in the wild-type and HR-deficient cells were lower or significantly lower than those in cells treated with C-ion beams. However, no differences were observed between the NHEJ-deficient cells with respect to the radiation type (Table 1 and Fig. 2B). In contrast, the β values did not differ with respect to the radiation type in any of the cell lines (Table 1).
FIG. 2.
Relative biological effectiveness (RBE) and α values. RBE values at 10% (RBE0.1) survival of protons and C ions and α values of all radiation types examined were calculated according to LQ model. Panel A: RBE0.1 values of protons (●) and C ions (■) in wild-type V79 and AA8; HR-deficient irs1 and irs1SF; and NHEJ-deficient XR1 and V3 cell lines. Panel B: α values of γ rays (▼), protons (●) and C ions (■) in wild-type V79 and AA8 cells, HR-deficient irs1 and irs1SF and NHEJ-deficient XR1 and V3 cells. Student’s t test: ##P < 0.01, #P <0.05 compared with wild-type and repair-deficient cells; **P < 0.01, *P < 0.05 compared with C ions. Error bars represent SD.
Histone γ-H2AX Nuclear Focus Formation and Dissolution
To determine how the patterns of DSB rejoining kinetics change with different radiation types, we examined the formation and dissolution of γ-H2AX foci in wild-type and repair-deficient cells at different time points after irradiation with 2 Gy of γ rays, protons and C ions. Histone γ-H2AX nuclear foci in the wild-type and repair-deficient cell were counted at 30 min and 3, 5, 7, 12 and 24 h after irradiation.
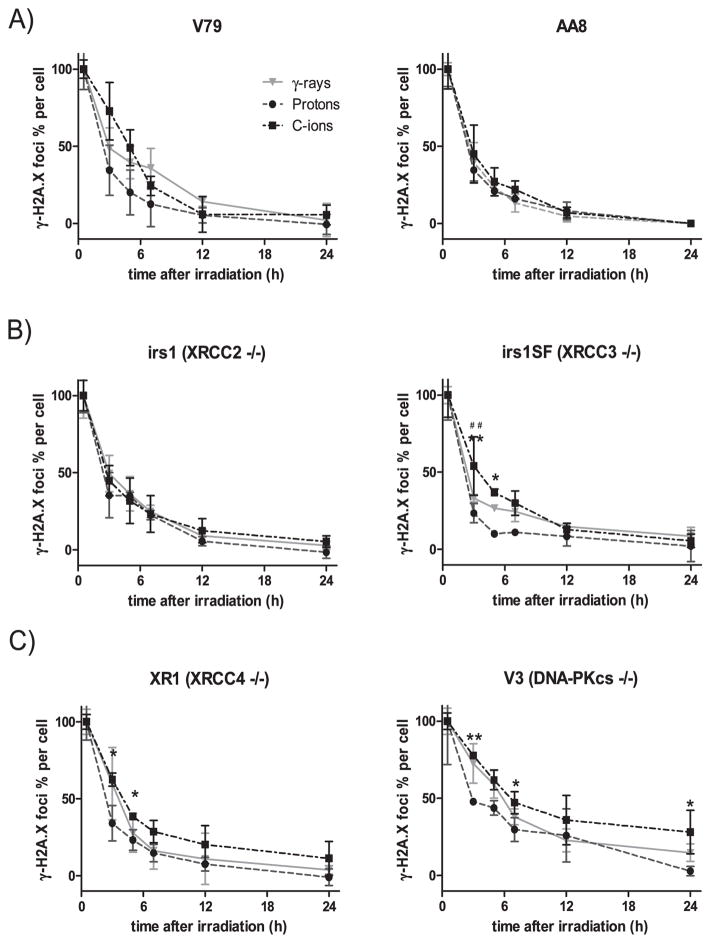
The average number of foci per cell before irradiation (background) was 3.76 ± 1.04, and the background values were subtracted from each obtained data point. The number of foci peaked at 30 min postirradiation (35.42 ± 4.66 after γ rays; 36.90 ± 6.06 after protons; 29.86 ± 4.43 after C ions) and then decreased gradually over time up to the 24 h point in all examined cell lines. The numbers of foci per cell at 30 min were normalized to 100% to compare the repair kinetics according to the number of γ-H2AX foci (Fig. 3). The percentages of γ-H2AX foci per cell did not significantly differ with respect to the three radiation types in the wild-type cell AA8 and the HR-deficient cell line irs1, however, they were higher, but insignificant, in wild-type V79 and significantly higher in the HR-deficient irs1SF cells for up to 5 h after C-ion irradiation (Fig. 3A, B) compared to protons. Furthermore, although the percentages of γ-H2AX foci per cell in the NHEJ-deficient cell lines did not significantly differ between γ rays and protons, the percentage of C-ion beam induced γ-H2AX foci remained higher especially at the early phases postirradiation (Fig. 3C). When comparing the wild-type and HR- and NHEJ-deficient cell lines over time after exposure to each radiation type, we found that the percentages of γ-H2AX foci decreased almost in the same manner in all cell lines, except for the NHEJ-deficient V3 cell line (Fig. 4B).
FIG. 3.
Quantification of γ-H2AX foci. Time response of γ-H2AX foci after irradiation with 2 Gy of γ rays (
 ), protons (●) and C ions (■). The percentage of foci per cell was plotted by normalizing the numbers at 30 min as 100% after subtracting the number of foci in 0 Gy irradiated cell. The γ-H2AX foci formation and dissolution in wild-type cell lines AA8 and V79 (panel A); HR-deficient cell lines irs1 and irs1SF (panel B); and NHEJ-deficient cell lines XR1 and V3 (panel C). Student’s t test: **P < 0.01, *P < 0.05 of C ions compared with protons; no significant differences were observed between C ions and γ rays. Error bars represent SD.
), protons (●) and C ions (■). The percentage of foci per cell was plotted by normalizing the numbers at 30 min as 100% after subtracting the number of foci in 0 Gy irradiated cell. The γ-H2AX foci formation and dissolution in wild-type cell lines AA8 and V79 (panel A); HR-deficient cell lines irs1 and irs1SF (panel B); and NHEJ-deficient cell lines XR1 and V3 (panel C). Student’s t test: **P < 0.01, *P < 0.05 of C ions compared with protons; no significant differences were observed between C ions and γ rays. Error bars represent SD.
Radiation-induced Chromosomal Aberrations
The wild-type (V79, AA8), HR- (irs1, irs1SF) and NHEJ-deficient (XR1, V3) cell lines were examined for various chromosomal aberrations induced by γ rays, protons and C ions. After each exposure, colcemid was added at 3 and 16 h and incubated for an additional 4 h before fixation. The numbers of chromosomal aberrations in nonirradiated cells were subtracted from those obtained at each time point. After irradiation, we compared the chromosomal aberrations at early (3 h) and late (16 h) time points in all cell lines and found a tendency towards an increase in aberrations at later time points in some cells, although no significant differences were observed. The incidence of chromosomal aberration was higher after C-ion irradiation than after other radiation types. The total number of chromosomal aberrations was significantly higher in the repair-deficient cells than in the wild-type cells (Fig. 4C).
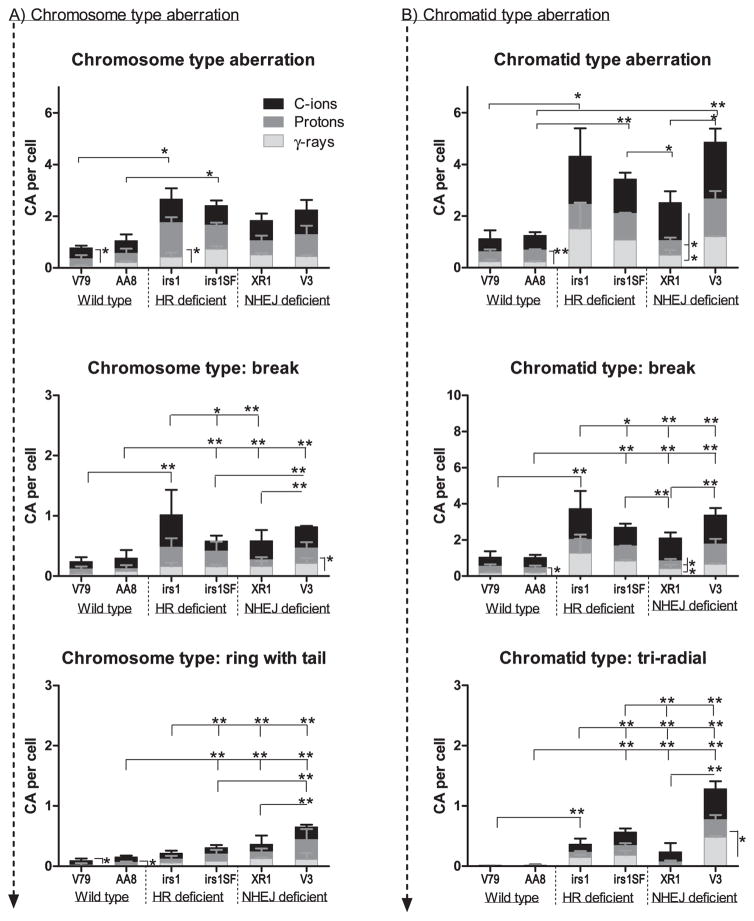
The types of aberrations observed in the wild-type and repair-deficient cells are shown in Fig. 5. Since there were no significant differences in types of chromosome aberration between early (3 h) and late (16 h) time points, only the results at 16 h were shown in Fig. 5. Both chromosome (Fig. 5A) and chromatid-type (Fig. 5B) aberrations were significantly higher in repair-deficient cells than in wild-type cells. The occurrences of chromosome type and chromatid-type aberrations did not significantly differ with respect to radiation types in any cell line. As to the chromatid-type aberrations, breaks and triradial types were more frequent in the HR- and NHEJ-deficient cells than in the wild-type cells (Fig. 5B). Among the repair-deficient cells, NHEJ-deficient XR1 (XRCC4−/−) cells showed significantly smaller numbers of chromatid-type aberrations, breaks and triradial types than other repair-deficient cell lines. Chromatid-type breaks occurred more frequently than other types of aberrations in all cell lines in all radiation types. In addition, for the chromosome type aberrations, the ring with tail was more frequent in the NHEJ-deficient cells, and breaks were more frequent in both HR- and NHEJ-deficient cells than in wild-type cells (Fig. 5A).
FIG. 5.
Chromosome and chromatid-type aberration at 16 h after irradiation. Panel A: The number of chromosome type aberrations and more frequently occurring chromosome type (break and ring with tail) aberrations per cell were plotted in each cell line after irradiation with γ rays (white column), protons (gray column) and C ions (black column). Panel B: The number of chromatid-type aberrations and more frequently occurring chromatid-type (break and triradial) aberrations per cell were plotted in each cell line after irradiation with γ rays (white column), protons (gray column) and C ions (black column). Student’s t test: **P < 0.01, *P < 0.05. The error bars represent SD.
DISCUSSION
Studies of how the NHEJ and HR pathways contribute to the repair of DSBs induced by different types of radiation are essential not only for understanding the responses of cancers to different radiotherapy types but also for developing molecularly targeted radiosensitizing therapies that incorporate different particle beams.
The choice of DSB repair pathways is a complex process that depends on many factors, including the DSB induction process (19, 34), repair protein proficiencies (10, 35), cell cycle phase (14, 15) and cell cycle checkpoint control (13, 36). Many pathways in addition to the major NHEJ and HR pathways have evolved to resolve DSBs, such as the alternative end-joining pathway, microhomology-mediated end-joining, synthesis-dependent strand annealing and single-strand annealing, and when one DSB repair pathway fails, others are activated (10, 11, 14). The occurrence and quality of radiation-induced DSBs are known to depend on the ionization density associated with the radiation (19, 34, 37), which potentially should exert a significant influence on the subsequent repair pathway choice. Therefore, in the current study we compared the contributions of the NHEJ and HR pathways in DSB repair in genetically established NHEJ- or HR-deficient cells after exposure to γ rays, protons and C ions.
NHEJ is a simple, rapid and efficient repair pathway that works throughout the cell cycle (14, 15, 38). DNA-PKcs is known to play a key role in this repair pathway and the phosphorylation status of DNA-PKcs itself indicates the level of kinase activity and the DNA end-binding ability, which probably determines the choice between the NHEJ and HR pathways (35, 36). In this study, we used two different NHEJ-deficient cell lines, V3 (DNA-PKcs−/−) and XR1 (XRCC4−/−), and our survival results showed that V3 was most sensitive to all radiation types (Fig. 1C).
The other NHEJ-deficient cell line, XR1, lacks the functional XRCC4 protein, which is known to play an important role in the ligation of broken ends via DNA ligase IV and XLF (39). In addition, XRCC4 is known to assist the recruitment of NHEJ-dependent DNA end-processing enzymes to the DSBs to facilitate the repair of specific breaks (40). However, XR1 cells exhibited less sensitivity to all radiation types when compared with V3 cells (Fig. 1C). This difference could indicate that although XRCC4 deficiency may induce incomplete NHEJ, this mutation is not as lethal as DNA-PKcs deficiency. Furthermore, other repair pathways may compensate for XRCC4 deficiency (10), while compensation for DNA-PKcs deficiency is not possible. These findings are compatible with other published studies in which DNA-PKcs was suggested to be the major protein involved in the repair pathway direction or selection (35, 41).
The HR-defective cell lines irs1 (XRCC2−/−) and irs1SF (XRCC3−/−) exhibited higher levels of sensitivity to all radiation types compared with the wild-type cells (Fig. 1A, B). XRCC2 and XRCC3 are paralogs of RAD51 that exist in two distinct complexes: one comprises RAD51B, RAD51C, RAD51D and XRCC2 (BCDX2) and the other comprises RAD51C and XRCC3 (CX3). The BCDX2 complex preferentially binds ssDNA and acts during the presynaptic phase of HR; the CX3 complex plays an essential role in the resolution of Holliday junctions created during HR (42, 43). From our survival results, XRCC3−/−irs1SF cells were 1.9-, 1.7- and 2.4-fold more sensitive than XRCC2−/− irs1 cells in response to γ-ray, proton and C-ion irradiation, respectively. Therefore it is possible that the contribution of XRCC3 is greater than XRCC2 in repair of DNA damage induced by all radiation types; in particular, its contribution may be greater after C-ion irradiation than other types of radiation.
The proton RBE0.1 values in the wild-type and NHEJ- and HR-deficient cells were not significantly different. In contrast, the C-ion RBE0.1 values in the NHEJ-deficient cells were significantly lower than those in the HR-deficient or wild-type cells. Furthermore, the C-ion and proton RBE0.1 values were nearly the same in the NHEJ-deficient cells, whereas the C-ion values were significantly higher than the proton values in the wild-type and HR-deficient cells (Table 1, Fig. 2A). As a result, the cytotoxicity ratios of C ions to protons were 10–20% higher in the NHEJ-deficient cells and 40–90% higher in the wild-type and HR-deficient cells. This could indicate that the repair of C-ion-induced DSBs requires a greater contribution from the HR pathway than of DSBs resulting from γ-ray or proton irradiation.
Furthermore, we compared the α values calculated according to the LQ model, which determines low-dose (up to 5 Gy) sensitivity and represents the contributions of single-event damage (44). The α values for C ions were significantly higher than those for γ rays or protons in the wild-type and irs1 (HR-deficient) cells. However, these values did not significantly differ in the NHEJ-deficient cells. In addition, α values of NHEJ-deficient cells were significantly higher than wild-type cells, while HR-deficient cells were not (Fig. 2B). These findings indicate that C-ion beams induce a greater amount of single-track DSBs than protons or γ rays “at the same dose”. This difference can be explained by the track structure of these types of radiation. That is, C ions have high-density ionizing cores that yield more single-track DSBs than sparsely ionizing protons or γ rays (45, 46). However, clinical C-ion beams are not absolutely pure, i.e., contamination by low-LET components due to fragmentation of the primary ions cannot be avoided (46), and the low-LET components including delta-ray ionization probably induce double-track DSBs at the penumbra region (45).
The fraction of residual γ-H2AX foci at 12 and 24 h after irradiation was higher in the NHEJ-deficient V3 and XR1 cells than in the wild-type and HR-deficient cells after C-ion irradiation (Fig. 4B), which may indicate that C-ion-induced DSBs are more irreparable than DSBs induced by γ-ray and proton irradiation. In particular, the residual fraction of γ-H2AX foci was significantly higher in the NHEJ-deficient V3 cells from 3–7 h after all types of irradiation than other cell lines (Fig. 4B), which is compatible with the clonogenic survival data shown in Fig. 4A. Mao et al. reported that the NHEJ pathway occurred within 30 min after irradiation, while the HR pathway required a much longer time (≥7 h) and that the efficiency of NHEJ was threefold higher than that of HR in actively cycling cells (38). In addition, the residual or persistent γ-H2AX foci at later time points may represent the chromatin alteration rather than unrepaired DSBs, and chromatin alterations are more likely to occur at hetero-chromatic regions repaired with slow kinetics (47). Therefore, the residual foci observed at later time points may indicate the occurrence of HR, and the HR pathway may have a greater role in C-ion-induced DSB repair.
The chromosomal aberrations after exposure to 1 Gy γ rays, protons and C ions were significantly higher in both the NHEJ and HR-deficient cells relative to the wild-type cells in response to all radiation types (Figs. 4C and 5). The high aberration levels in the HR-deficient cells might result from the increased rate of DSB repair via the error-prone NHEJ pathway. Furthermore, the differences in aberration levels between the NHEJ-deficient XR1 and V3 cell lines could be explained as follows: error-free HR compensated for NHEJ impairment due to the loss of XRCC4 (XR1) but not DNA-PKcs (V3). However, definitive reasons to explain these observations remain unidentified. Overall, the chromosomal aberration level did not significantly differ in response to γ rays and protons, whereas C-ion radiation induced increased aberration levels in all cell lines. This may indicate that although both NHEJ and HR pathways are essential, C-ion-induced DNA damage is not easily repair correctly.
Recently, Grosse et al. compared the cytotoxicities of photons and protons in the same cell lines and reported that proton-induced DSBs preferentially require the HR repair pathway (48). However, we did not observe a significant difference after γ-ray and proton irradiation. This discrepancy was likely due to differences in the colony formation assay method. Grosse et al. incubated irradiated cells for 20 h before seeding. Barendsen noted that single-track damage was not repaired by delayed plating and that single-track lethal damage encompassed two types of damage: one component is not repaired by delayed plating and is very strongly dependent on LET, and the other component comprises potentially lethal damage that is weakly dependent on LET (49). Thus, Gross et al. may have evaluated the contributions of HR and NHEJ to the repair of single-event type damage that could not be repaired by delayed plating (44, 49). In contrast, our colony formation results represent all types of damage that lead to reproductive death, including single-event type, sublethal and potentially lethal damage, suggesting that the NHEJ pathway plays a greater role than the HR pathway in DNA damage repair caused by exposure not only to γ rays but also to protons. In addition, recent published studies have shown that γ-ray-induced DSB rejoining is strictly dependent on the NHEJ repair system integrity and the NHEJ plays a more important role in repair after C-ion exposure (14, 50). Our results, which are compatible with that earlier report, suggest that NHEJ is the major pathway and that among many factors, DNA-PKcs plays the most important role in regulating the process of repairing DSBs induced by exposure to protons as well as γ rays.
In conclusion, the DNA-PKcs-dependent NHEJ pathway plays an important role in repairing DSBs induced by both clinical proton and C-ion beams. Furthermore, our results suggest that the HR pathway is more involved in the repair of DSBs induced by C-ion beams than those induced by proton beams or γ rays.
Acknowledgments
We are indebted to Ms. Junko Zenkoh and Ms. Yoko Mori, as well as all the members of the PMRC at the University of Tsukuba for their competent technical assistance. We are also grateful to the members of the HIMAC facility at the National Institute of Radiological Sciences for their valuable help in our experiments. In addition, we thank Mr. Takamitsu Kato for providing the cell line and for his valuable advice. This work was supported in part by a Grant-in-Aid (24390287) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and by the National Aeronautics and Space Association (NNX13AD57G to AA and DJC). Portions of this work were approved and performed under the Research Project with Heavy Ions at NIRS-HIMAC.
References
- 1.Ando K, Koike S, Kawachi K, Hiraoka T, Ohara H, Yokota M, et al. Relative biological effectiveness of the therapeutic proton beams at NIRS and Tsukuba University. Nihon Igaku Hoshasen Gakkai Zasshi. 1985;45:531–5. [PubMed] [Google Scholar]
- 2.Belli M, Cera F, Cherubini R, Haque AM, Ianzini F, Moschini G, et al. Inactivation and mutation induction in V79 cells by low energy protons: re-evaluation of the results at the LNL facility. Int J Radiat Biol. 1993;63:331–7. doi: 10.1080/09553009314550441. [DOI] [PubMed] [Google Scholar]
- 3.Di Pietro C, Piro S, Tabbi G, Ragusa M, Di Pietro V, Zimmitti V, et al. Cellular and molecular effects of protons: Apoptosis induction and potential implications for cancer therapy. Apoptosis. 2006;11:57–66. doi: 10.1007/s10495-005-3346-1. [DOI] [PubMed] [Google Scholar]
- 4.Gerelchuluun A, Hong Z, Sun L, Suzuki K, Terunuma T, Yasuoka K, et al. Induction of in situ DNA double-strand breaks and apoptosis by 200 MeV protons and 10 MV X-rays in human tumour cell lines. Int J Radiat Biol. 2011;87:57–70. doi: 10.3109/09553002.2010.518201. [DOI] [PubMed] [Google Scholar]
- 5.Hong Z, Kase Y, Moritake T, Gerelchuluun A, Sun L, Suzuki K, et al. Lineal energy-based evaluation of oxidative DNA damage induced by proton beams and X-rays. Int J Radiat Biol. 2013;89:36–43. doi: 10.3109/09553002.2012.715791. [DOI] [PubMed] [Google Scholar]
- 6.Kagawa K, Murakami M, Hishikawa Y, Abe M, Akagi T, Yanou T, et al. Preclinical biological assessment of proton and carbon ion beams at Hyogo Ion Beam Medical Center. Int J Radiat Oncol Biol Phys. 2002;54:928–38. doi: 10.1016/s0360-3016(02)02949-8. [DOI] [PubMed] [Google Scholar]
- 7.Koike S, Ando K, Oohira C, Fukawa T, Lee R, Takai N, et al. Relative biological effectiveness of 290 MeV/u carbon ions for the growth delay of a radioresistant murine fibrosarcoma. J Radiat Res. 2002;43:247–55. doi: 10.1269/jrr.43.247. [DOI] [PubMed] [Google Scholar]
- 8.Davis AJ, Chen BP, Chen DJ. DNA-PK: a dynamic enzyme in a versatile DSB repair pathway. DNA Repair (Amst) 2014;17:21–9. doi: 10.1016/j.dnarep.2014.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langerak P, Mejia-Ramirez E, Limbo O, Russell P. Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks. PLoS Genet. 2011;7:e1002271. doi: 10.1371/journal.pgen.1002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001;15:3237–42. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reynolds P, Anderson JA, Harper JV, Hill MA, Botchway SW, Parker AW, et al. The dynamics of Ku70/80 and DNA-PKcs at DSBs induced by ionizing radiation is dependent on the complexity of damage. Nucleic Acids Research. 2012;40:10821–31. doi: 10.1093/nar/gks879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krejci L, Altmannova V, Spirek M, Zhao X. Homologous recombination and its regulation. Nucleic Acids Res. 2012;40:5795–818. doi: 10.1093/nar/gks270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mao Z, Bozzella M, Seluanov A, Gorbunova V. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle. 2008;7:2902–6. doi: 10.4161/cc.7.18.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bee L, Fabris S, Cherubini R, Mognato M, Celotti L. The efficiency of homologous recombination and non-homologous end joining systems in repairing double-strand breaks during cell cycle progression. PLoS One. 2013;8:e69061. doi: 10.1371/journal.pone.0069061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol. 2003;23:5706–15. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whitmore GF, Varghese AJ, Gulyas S. Cell cycle responses of two X-ray sensitive mutants defective in DNA repair. Int J Radiat Biol. 1989;56:657–65. doi: 10.1080/09553008914551881. [DOI] [PubMed] [Google Scholar]
- 17.Yajima H, Fujisawa H, Nakajima NI, Hirakawa H, Jeggo PA, Okayasu R, et al. The complexity of DNA double strand breaks is a critical factor enhancing end-resection. DNA Repair. 2013;12:936–46. doi: 10.1016/j.dnarep.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 18.Asaithamby A, Chen DJ. Mechanism of cluster DNA damage repair in response to high-atomic number and energy particles radiation. Mutat Res. 2011;711:87–99. doi: 10.1016/j.mrfmmm.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hada M, Sutherland BM. Spectrum of complex DNA damages depends on the incident radiation. Radiat Res. 2006;165:223–30. doi: 10.1667/rr3498.1. [DOI] [PubMed] [Google Scholar]
- 20.Wang H, Wang X, Zhang P, Wang Y. The Ku-dependent non-homologous end-joining but not other repair pathway is inhibited by high linear energy transfer ionizing radiation. DNA Repair (Amst) 2008;7:725–33. doi: 10.1016/j.dnarep.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 21.Fuller LF, Painter RB. A Chinese hamster ovary cell line hypersensitive to ionizing radiation and deficient in repair replication. Mutat Res. 1988;193:109–21. doi: 10.1016/0167-8817(88)90041-7. [DOI] [PubMed] [Google Scholar]
- 22.Chu EH, Brimer P, Jacobson KB, Merriam EV. Mammalian cell genetics. I. Selection and characterization of mutations auxotrophic for L-glutamine or resistant to 8-azaguanine in Chinese hamster cells in vitro. Genetics. 1969;62:359–77. doi: 10.1093/genetics/62.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakaguchi DS, Moeller JF, Coffman CR, Gallenson N, Harris WA. Growth cone interactions with a glial cell line from embryonic Xenopus retina. Dev Biol. 1989;134:158–74. doi: 10.1016/0012-1606(89)90086-9. [DOI] [PubMed] [Google Scholar]
- 24.Tebbs RS, Zhao Y, Tucker JD, Scheerer JB, Siciliano MJ, Hwang M, et al. Correction of chromosomal instability and sensitivity to diverse mutagens by a cloned cDNA of the XRCC3 DNA repair gene. Proc Natl Acad Sci U S A. 1995;92:6354–8. doi: 10.1073/pnas.92.14.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones NJ, Cox R, Thacker J. Isolation and cross-sensitivity of X-ray-sensitive mutants of V79-4 hamster cells. Mutat Res. 1987;183:279–86. doi: 10.1016/0167-8817(87)90011-3. [DOI] [PubMed] [Google Scholar]
- 26.Nohtomi A, Sakae T, Tsunashima Y, Kohno R. Dosimetry of pulsed clinical proton beams by a small ionization. Med Phys. 2001;28:1431–5. doi: 10.1118/1.1380435. [DOI] [PubMed] [Google Scholar]
- 27.Wilkens JJ, Oelfke U. Analytical linear energy transfer calculations for proton therapy. Med Phys. 2003;30:806–15. doi: 10.1118/1.1567852. [DOI] [PubMed] [Google Scholar]
- 28.Stopping powers and ranges for protons and alpha particles (ICRU Report 49) Bethesda: International Commission on Radiation Units and Measurements; 1993. [Google Scholar]
- 29.Fukumura A, Hiraoka T, Omata K, Takeshita M, Kawachi K, Kanai T, et al. Carbon beam dosimetry intercomparison at HIMAC. Phys Med Biol. 1998;43:3459–63. doi: 10.1088/0031-9155/43/12/005. [DOI] [PubMed] [Google Scholar]
- 30.Kanai T, Endo M, Minohara S, Miyahara N, Koyama-ito H, Tomura H, et al. Biophysical characteristics of HIMAC clinical irradiation system for heavy-ion radiation therapy. Int J Radiat Oncol Biol Phys. 1999;44:201–10. doi: 10.1016/s0360-3016(98)00544-6. [DOI] [PubMed] [Google Scholar]
- 31.Nagasawa H, Brogan JR, Peng Y, Little JB, Bedford JS. Some unsolved problems and unresolved issues in radiation cytogenetics: a review and new data on roles of homologous recombination and non-homologous end joining. Mutat Res. 2010;701:12–22. doi: 10.1016/j.mrgentox.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 32.Cornforth MN, Bedford JS. X-ray-induced breakage and rejoining of human interphase chromosomes. Science. 1983;222:1141–3. doi: 10.1126/science.6648528. [DOI] [PubMed] [Google Scholar]
- 33.Savage JRK. Classification and relationships of induced chromosomal structural-changes. J Med Genet. 1976;13:103–22. doi: 10.1136/jmg.13.2.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Belli M, Cherubini R, Dalla Vecchia M, Dini V, Esposito G, Moschini G, et al. DNA fragmentation in mammalian cells exposed to various light ions. Adv Space Res. 2001;27:393–9. doi: 10.1016/s0273-1177(01)00007-2. [DOI] [PubMed] [Google Scholar]
- 35.Neal JA, Dang V, Douglas P, Wold MS, Lees-Miller SP, Meek K. Inhibition of homologous recombination by DNA-dependent protein kinase requires kinase activity, is titratable, and is modulated by autophosphorylation. Mol Cell Biol. 2011;31:1719–33. doi: 10.1128/MCB.01298-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shrivastav M, Miller CA, De Haro LP, Durant ST, Chen BP, Chen DJ, et al. DNA-PKcs and ATM co-regulate DNA double-strand break repair. DNA Repair (Amst) 2009;8:920–9. doi: 10.1016/j.dnarep.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eccles LJ, O’Neill P, Lomax ME. Delayed repair of radiation induced clustered DNA damage: friend or foe? Mutat Res. 2011;711:134–41. doi: 10.1016/j.mrfmmm.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mao Z, Bozzella M, Seluanov A, Gorbunova V. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair (Amst) 2008;7:1765–71. doi: 10.1016/j.dnarep.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grawunder U, Wilm M, Wu XT, Kulesza P, Wilson TE, Mann M, et al. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388:492–5. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- 40.Kusumoto R, Dawut L, Marchetti C, Lee JW, Vindigni A, Ramsden D, et al. Werner protein cooperates with the XRCC4-DNA ligase IV complex in end-processing. Biochemistry. 2008;47:7548–56. doi: 10.1021/bi702325t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hammel M, Yu Y, Mahaney BL, Cai B, Ye R, Phipps BM, et al. Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. J Biol Chem. 2010;285:1414–23. doi: 10.1074/jbc.M109.065615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, Tarsounas M, O’Regan P, West SC. Role of RAD51C and XRCC3 in genetic recombination and DNA repair. J Biol Chem. 2007;282:1973–9. doi: 10.1074/jbc.M609066200. [DOI] [PubMed] [Google Scholar]
- 43.Masson JY, Tarsounas MC, Stasiak AZ, Stasiak A, Shah R, McIlwraith MJ, et al. Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev. 2001;15:3296–307. doi: 10.1101/gad.947001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Franken NAP, Oei AL, Kok HP, Rodermond HM, Sminia P, Crezee J, et al. Cell survival and radiosensitisation: Modulation of the linear and quadratic parameters of the LQ model (Review) Int J Oncol. 2013;42:1501–15. doi: 10.3892/ijo.2013.1857. [DOI] [PubMed] [Google Scholar]
- 45.Nakajima NI, Brunton H, Watanabe R, Shrikhande A, Hirayama R, Matsufuji N, et al. Visualisation of gamma H2AX foci caused by heavy ion particle traversal; distinction between core track versus non-track damage. Plos One. 2013:8. doi: 10.1371/journal.pone.0070107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hirayama R, Ito A, Tomita M, Tsukada T, Yatagai F, Noguchi M, et al. Contributions of direct and indirect actions in cell killing by high-LET radiations. Radiat Res. 2009;171:212–8. doi: 10.1667/RR1490.1. [DOI] [PubMed] [Google Scholar]
- 47.Costes SV, Chiolo I, Pluth JM, Barcellos-Hoff MH, Jakob B. Spatiotemporal characterization of ionizing radiation induced DNA damage foci and their relation to chromatin organization. Mutat Res. 2010;704:78–87. doi: 10.1016/j.mrrev.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grosse N, Fontana AO, Hug EB, Lomax A, Coray A, Augsburger M, et al. Deficiency in homologous recombination renders Mammalian cells more sensitive to proton versus photon irradiation. Int J Radiat Oncol Biol Phys. 2014;88:175–81. doi: 10.1016/j.ijrobp.2013.09.041. [DOI] [PubMed] [Google Scholar]
- 49.Barendsen GW. The relationships between Rbe and Let for different types of lethal damage in mammalian-cells – biophysical and molecular mechanisms. Radiat Res. 1994;139:257–70. [PubMed] [Google Scholar]
- 50.Takahashi A, Kubo M, Ma H, Nakagawa A, Yoshida Y, Isono M, et al. Non-homologous end-joining repair plays a more important role than homologous recombination repair in defining radiosensitivity after exposure to high-LET radiation. Radiat Res. 2014;182:338–44. doi: 10.1667/RR13782.1. [DOI] [PubMed] [Google Scholar]