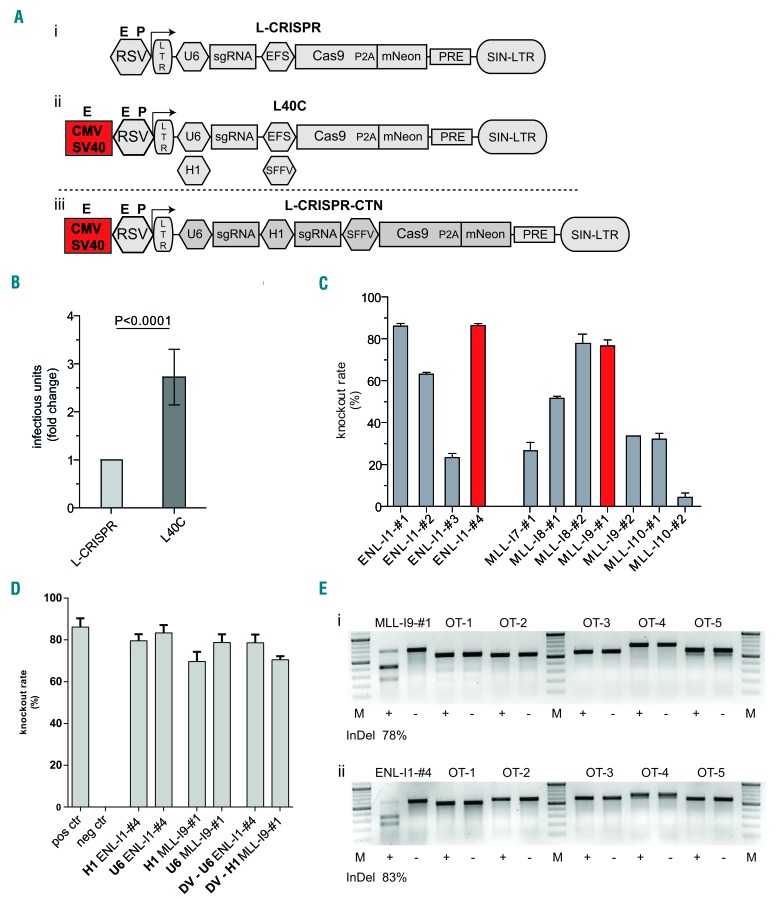
Figure 1.
An improved lentiviral vector system for generation of CRISPR-Cas9-induced chromosomal rearrangements. (A) Schematic presentation of lentiviral vector architecture including genomic RNA-generating promoter assembly: published architecture (L-CRISPR) (i), improved architecture with cytomegalovirus enhancer (CMV) and simian virus 40 enhancer (SV40) and exchangeability of the hU6 promoter for a H1 promoter (L40C) (ii), and lentiviral vector for dual sgRNA delivery (L-CRISPR-CTN (iii). (B) Analysis of viral titers in three independent cell lines with two different sgRNAs and three replicates each. (C) Knock-out efficacies (fluorescence reporter assay) of sgRNAs targeting intronic sequences of ENL and MLL. Selected sgRNAs are marked. (D) Knock-out efficacies of selected sgRNAs expressed from a human U6 or H1 promoter, as indicated. Knock-out efficacies of selected sgRNAs in L-CRISPR-CTN dual sgRNA configuration (DV) (neg ctrl= anti-luciferase (Luc) sgRNA, pos ctrl= Tet2 sgRNA). (E) T7-endonuclease-I assay for on-target sites and the top five predicted off-target sites in HEL cells (OT-1-OT-5). Indel frequencies at endogenous loci are indicated below. Analysis in cells transduced with targeting (+) and Luc (-) sgRNAs: MLL-I9-#1(i) and ENL-I1-#4 (ii).