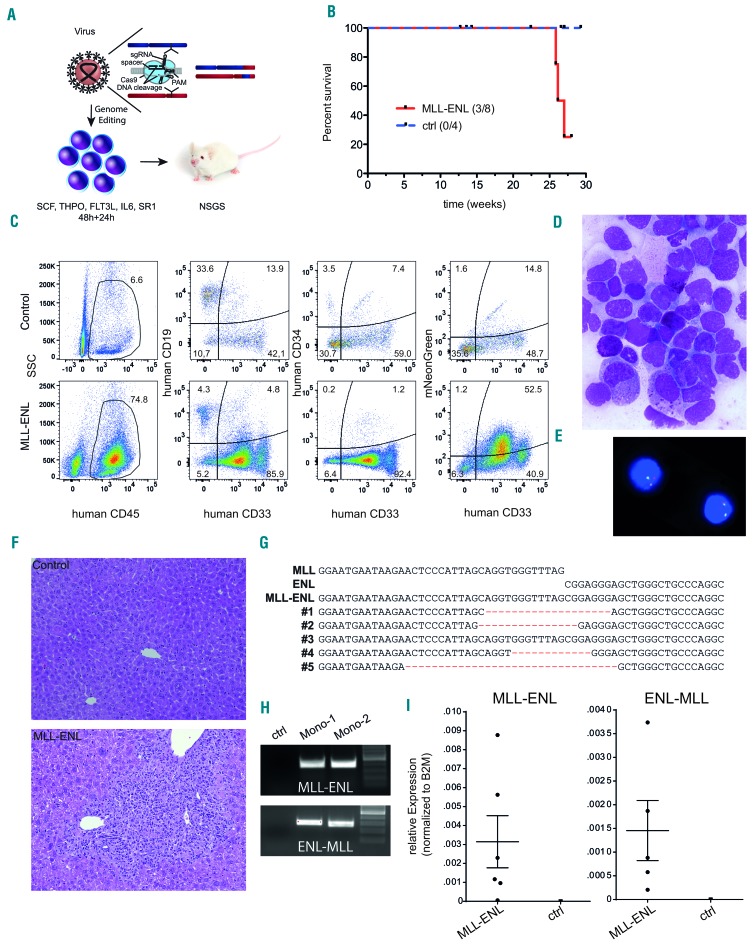
Figure 3.
CRISPR-Cas9-induced MLL-ENL rearrangements are leukemogenic in a CD34+ hematopoietic stem and progenitor cell xenotransplantation model. (A) Schematic depiction of the xenotransplantation model. (B) Survival of mice transplanted with L-CRISPR-CTN-transduced CD34+ HSPC. Mice that succumbed to non-hematopoietic disease were censored and are indicated (ticked). (C) Flow cytometric analysis of one mouse with hematopoietic disease compared to a control, with markers as indicated (ctrl = MLL-I9-#1 + Luc sgRNAs). (D) Bone marrow (BM) cytospin analysis of a diseased mouse (MGG, 1000X). (E) Detection of an MLL translocation with fluorescence in situ hybridization on interphase nuclei (Vysis LSI MLL probe; Abbott Laboratories) in BM cells from one diseased mouse. (F) Histopathological analysis of liver tissue from a healthy control mouse (ctrl = MLL-I9-#1 + Luc sgRNA) (top) and a mouse with monocytic leukemia-like disease transplanted with L-CRISPR-CTN(11;19)-containing CD34+ HSPC (bottom) (HE, 100x). (G) Alignment of Sanger sequencing-derived genomic t(11;19)/MLL-ENL breakpoints of mice with a detectable MLL-ENL breakpoint. (H) MLL-ENL and ENL-MLL genomic breakpoints detected in the BM of two mice with a monocytic leukemia-like disease. (I) Expression of the MLL-ENL (left) and reciprocal ENL-MLL (right) fusion genes, measured by quantitative polymerase chian reaction from the BM of mice with a detectable MLL-ENL breakpoint compared to control mice (MLL-I9-#1 + Luc sgRNAs).