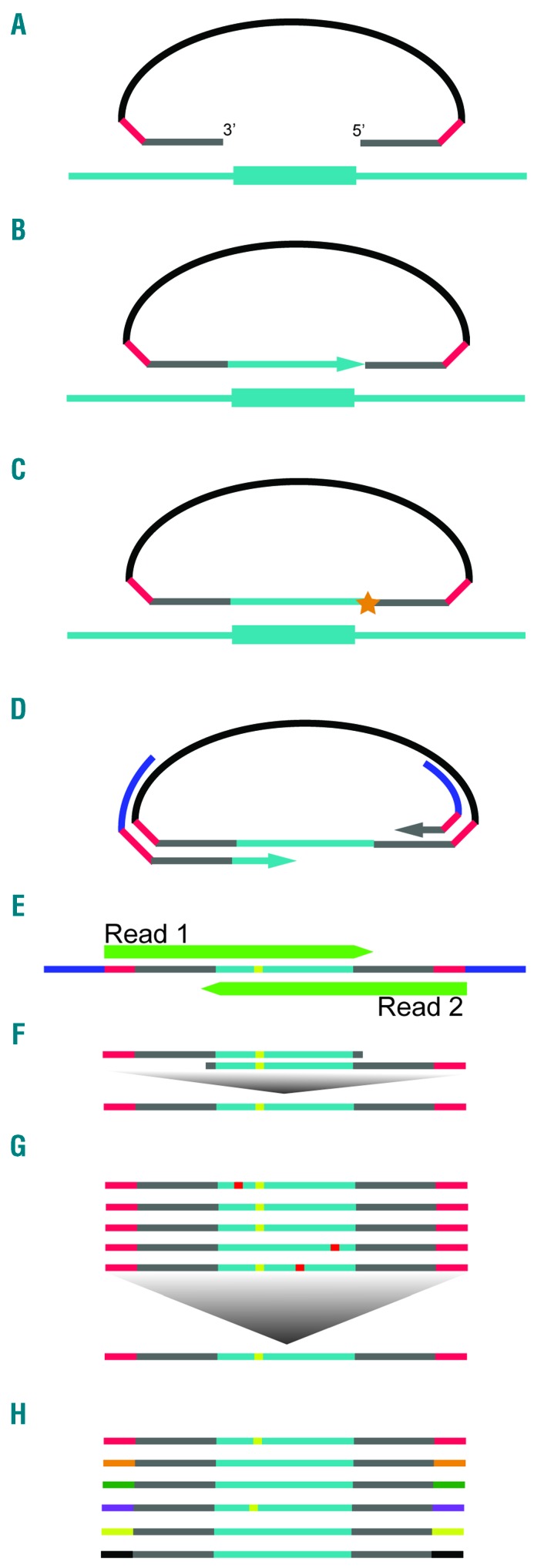
Figure 1.
Overview of smMIP capture. (A) smMIPs are single-stranded (ss)DNA oligonucleotides with domains at both ends that are complimentary to genomic targets of interest (gray), each flanked by a 4bp fully degenerate UMID sequence (red), totaling an 8bp molecular tag, and linked by a backbone sequence common to all probes (black). After hybridization of probes to DNA, (B) DNA polymerase copies targeted genomic DNA by extension of the free probe arm and (C) the smMIP is joined into a covalently sealed circle by the action of DNA ligase. After exonuclease digestion of unbound probes and free genomic DNA, (D) PCR primed against the defined smMIP backbone is performed to amplify successful capture events and their identifying UMIDs. (E) Paired-end, high-throughput sequencing is performed. (F) Derived paired-end reads are merged into a single, contiguous read, providing inherent intra-read error correction. (G) A consensus sequence is generated out of all reads sharing a common UMID. Low prevalence, random errors are canceled out. (H) Extremely low variant allele frequencies can be accurately quantified by assaying the fraction of error-corrected reads bearing a mutation of interest out of the larger population of read groups derived from unique UMID-tagged smMIPs.