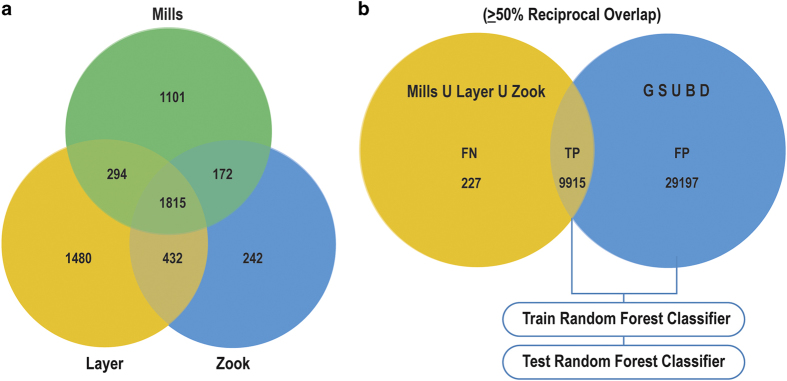
Figure 2.
Development of a DSV prediction classifier with reference data. (a) Concordance of confirmed DSVs among three published NA12878 reference DSV sets (Mills et al.,40 Layer et al.41and Zook et al.42). DSVs were considered concordant if their chromosomal coordinates had a reciprocal overlap of at least 50%. About 33% of DSV calls were common to all three sets; 49% were common to more than two sets. Rates of concordance were lower if the overlap requirement was increased. (b) Process employed to develop a DSV prediction classifier. FN, false negative; FP, false positive; TP, true positive. Numbers shown represent a 50% reciprocal intersection between the union of DSVs from Mills et al.,13 Layer et al.41 and Zook et al.,42 and the union of GS and BD calls for NA12878 technical replicates. TPs were defined as calls which had a ⩾50% reciprocal overlap between Mills U Layer U Zook and GS U BD. FNs were defined as Mills_Layer_Zook DSVs that were not found to have a ⩾50% reciprocal overlap with GS U BD calls. FPs were defined as GS U BD calls not found to have a 50% reciprocal overlap with Mills_Layer_Zook DSVs.